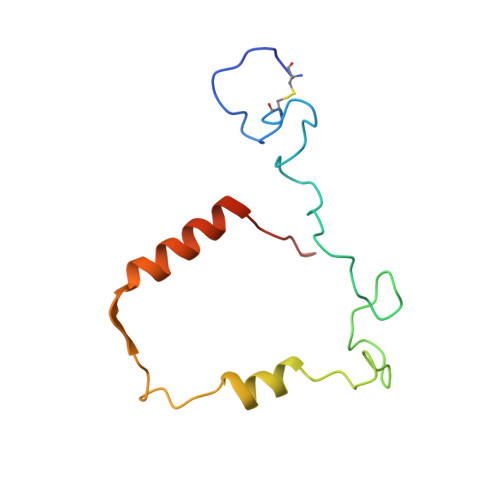
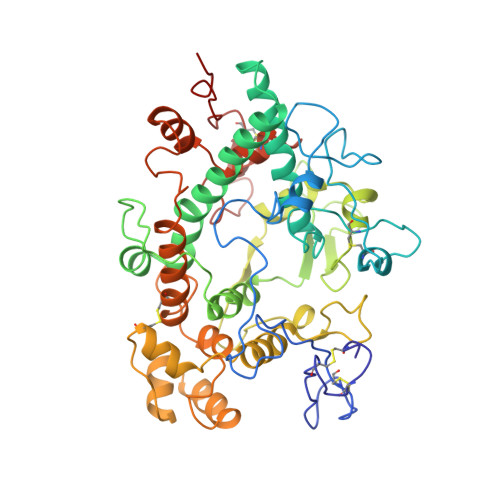
Structure of the green heme in myeloperoxidase.
Fenna, R., Zeng, J., Davey, C.(1995) Arch Biochem Biophys 316: 653-656
- PubMed: 7840679
- DOI: https://doi.org/10.1006/abbi.1995.1086
- Primary Citation of Related Structures:
1MHL - PubMed Abstract:
A 3-A-resolution X-ray crystal structure of canine myeloperoxidase has previously revealed the overall structure of the molecule, including the polypeptide backbone conformation, but did not provide an unambiguous structure for the covalently bound heme. A higher resolution (2.28 A) X-ray crystal structure of human myeloperoxidase has now shown that the heme is a novel derivative of protoporphyrin IX in which three ring substituents form covalent bonds with amino acid side chains in the protein. Modified methyl groups on pyrrole rings A and C form ester linkages with glutamate 242 and aspartate 94, while a covalent bond between the vinyl group on ring A and the sulfur atom of methionine 243 results in a sulfonium ion linkage. The heme tetrapyrrole ring also shows considerable distortion from the planar conformation seen in most heme-containing proteins. The observed bending appears to result from these covalent bonds between diametrically opposed pyrrole rings A and C and the protein. Sequence comparisons suggest that the two ester linkages to the heme may also occur in other homologous mammalian peroxidases, but that the sulfonium ion linkage may be a unique feature of myeloperoxidase.
- Department of Biochemistry and Molecular Biology, University of Miami, Florida 33101.
Organizational Affiliation: