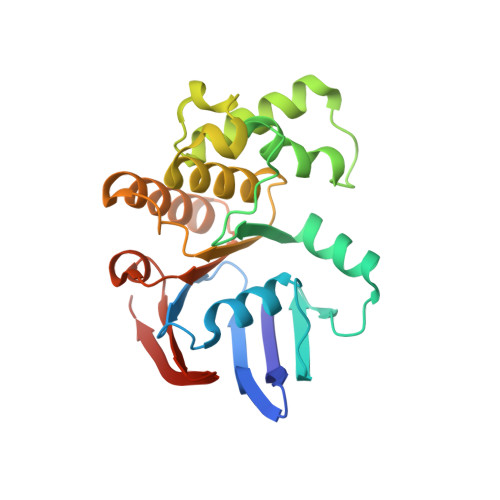
ATP binding to the motor domain from an ABC transporter drives formation of a nucleotide sandwich dimer.
Smith, P.C., Karpowich, N., Millen, L., Moody, J.E., Rosen, J., Thomas, P.J., Hunt, J.F.(2002) Mol Cell 10: 139-149
- PubMed: 12150914
- DOI: https://doi.org/10.1016/s1097-2765(02)00576-2
- Primary Citation of Related Structures:
1L2T - PubMed Abstract:
It has been proposed that the reaction cycle of ATP binding cassette (ABC) transporters is driven by dimerization of their ABC motor domains upon binding ATP at their mutual interface. However, no such ATP sandwich complex has been observed for an ABC from an ABC transporter. In this paper, we report the crystal structure of a stable dimer formed by the E171Q mutant of the MJ0796 ABC, which is hydrolytically inactive due to mutation of the catalytic base. The structure shows a symmetrical dimer in which two ATP molecules are each sandwiched between the Walker A motif in one subunit and the LSGGQ signature motif in the other subunit. These results establish the stereochemical basis of the power stroke of ABC transporter pumps.
- Department of Biological Sciences, 702A Fairchild Center, MC2434, Columbia University, New York, New York 10027, USA.
Organizational Affiliation: