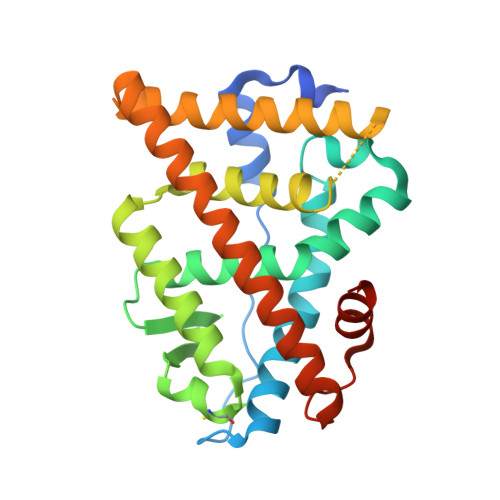
Structural characterization of a subtype-selective ligand reveals a novel mode of estrogen receptor antagonism.
Shiau, A.K., Barstad, D., Radek, J.T., Meyers, M.J., Nettles, K.W., Katzenellenbogen, B.S., Katzenellenbogen, J.A., Agard, D.A., Greene, G.L.(2002) Nat Struct Biol 9: 359-364
- PubMed: 11953755
- DOI: https://doi.org/10.1038/nsb787
- Primary Citation of Related Structures:
1L2I, 1L2J - PubMed Abstract:
The R,R enantiomer of 5,11-cis-diethyl-5,6,11,12-tetrahydrochrysene-2,8-diol (THC) exerts opposite effects on the transcriptional activity of the two estrogen receptor (ER) subtypes, ER alpha and ER beta. THC acts as an ER alpha agonist and as an ER beta antagonist. We have determined the crystal structures of the ER alpha ligand binding domain (LBD) bound to both THC and a fragment of the transcriptional coactivator GRIP1, and the ER beta LBD bound to THC. THC stabilizes a conformation of the ER alpha LBD that permits coactivator association and a conformation of the ER beta LBD that prevents coactivator association. A comparison of the two structures, taken together with functional data, reveals that THC does not act on ER beta through the same mechanisms used by other known ER antagonists. Instead, THC antagonizes ER beta through a novel mechanism we term 'passive antagonism'.
- The Howard Hughes Medical Institute and Department of Biochemistry and Biophysics, University of California, San Francisco, California 94143, USA.
Organizational Affiliation: