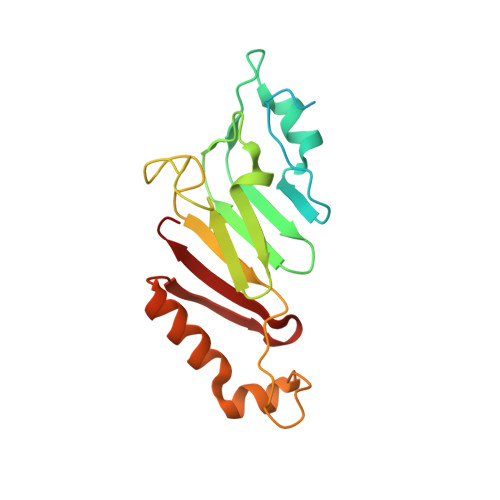
The three-dimensional structure of the autoproteolytic, nuclear pore-targeting domain of the human nucleoporin Nup98.
Hodel, A.E., Hodel, M.R., Griffis, E.R., Hennig, K.A., Ratner, G.A., Xu, S., Powers, M.A.(2002) Mol Cell 10: 347-358
- PubMed: 12191480
- DOI: https://doi.org/10.1016/s1097-2765(02)00589-0
- Primary Citation of Related Structures:
1KO6 - PubMed Abstract:
Nup98 is a component of the nuclear pore that plays its primary role in the export of RNAs. Nup98 is expressed in two forms, derived from alternate mRNA splicing. Both forms are processed into two peptides through autoproteolysis mediated by the C-terminal domain of hNup98. The three-dimensional structure of the C-terminal domain reveals a novel protein fold, and thus a new class of autocatalytic proteases. The structure further reveals that the suggested nucleoporin RNA binding motif is unlikely to bind to RNA. The C terminus also contains sequences that target hNup98 to the nuclear pore complex. Noncovalent interactions between the C-terminal domain and the cleaved peptide tail are visible and suggest a model for cleavage-dependent targeting of hNup98 to the nuclear pore.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, GA 30322, USA. ahodel@emory.edu
Organizational Affiliation: