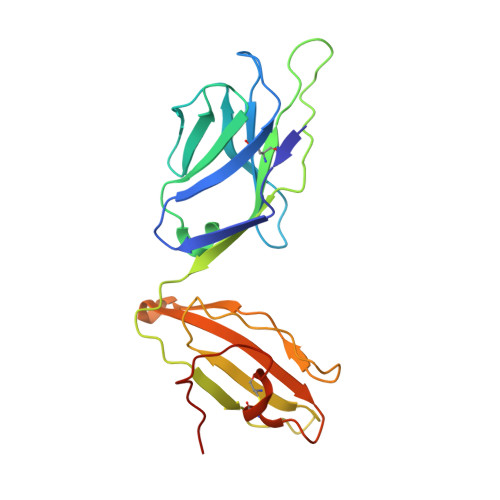
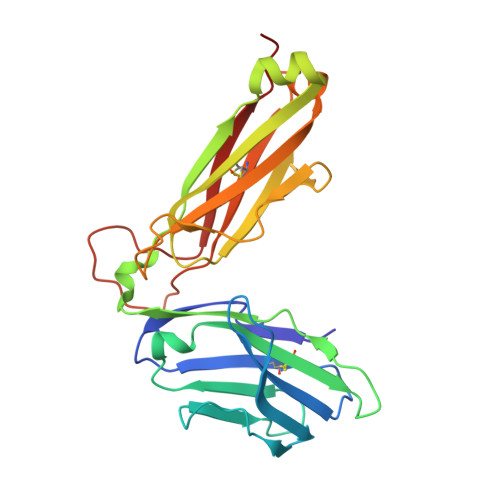
The 1.5 A crystal structure of a highly selected antiviral T cell receptor provides evidence for a structural basis of immunodominance
Kjer-Nielsen, L., Clements, C.S., Brooks, A.G., Purcell, A.W., McCluskey, J., Rossjohn, J.(2002) Structure 10: 1521-1532
- PubMed: 12429093
- DOI: https://doi.org/10.1016/s0969-2126(02)00878-x
- Primary Citation of Related Structures:
1KGC - PubMed Abstract:
Despite a potential repertoire of >10(15) alphabeta T cell receptors (TcR), the HLA B8-restricted cytolytic T cell response to a latent antigen of Epstein-Barr virus (EBV) is strikingly limited in the TcR sequences that are selected. Even in unrelated individuals this response is dominated by a single highly restricted TcR clonotype that selects identical combinations of hypervariable Valpha, Vbeta, D, J, and N region genes. We have determined the 1.5 A crystal structure of this "public" TcR, revealing that five of the six hypervariable loops adopt novel conformations providing a unique combining site that contains a deep pocket predicted to overlay the HLA B8-peptide complex. The findings suggest a structural basis for the immunodominance of this clonotype in the immune response to EBV.
- Department of Microbiology and Immunology, University of Melbourne, Parkville, Victoria, Australia.
Organizational Affiliation: