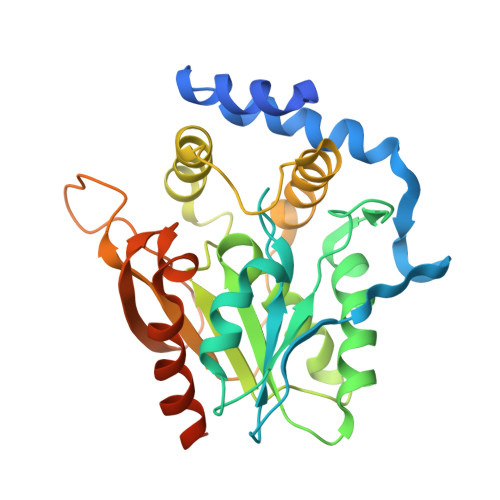
Crystal structure of the macrocycle-forming thioesterase domain of the erythromycin polyketide synthase: versatility from a unique substrate channel.
Tsai, S.C., Miercke, L.J., Krucinski, J., Gokhale, R., Chen, J.C., Foster, P.G., Cane, D.E., Khosla, C., Stroud, R.M.(2001) Proc Natl Acad Sci U S A 98: 14808-14813
- PubMed: 11752428
- DOI: https://doi.org/10.1073/pnas.011399198
- Primary Citation of Related Structures:
1KEZ - PubMed Abstract:
As the first structural elucidation of a modular polyketide synthase (PKS) domain, the crystal structure of the macrocycle-forming thioesterase (TE) domain from the 6-deoxyerythronolide B synthase (DEBS) was solved by a combination of multiple isomorphous replacement and multiwavelength anomalous dispersion and refined to an R factor of 24.1% to 2.8-A resolution. Its overall tertiary architecture belongs to the alpha/beta-hydrolase family, with two unusual features unprecedented in this family: a hydrophobic leucine-rich dimer interface and a substrate channel that passes through the entire protein. The active site triad, comprised of Asp-169, His-259, and Ser-142, is located in the middle of the substrate channel, suggesting the passage of the substrate through the protein. Modeling indicates that the active site can accommodate and orient the 6-deoxyerythronolide B precursor uniquely, while at the same time shielding the active site from external water and catalyzing cyclization by macrolactone formation. The geometry and organization of functional groups explain the observed substrate specificity of this TE and offer strategies for engineering macrocycle biosynthesis. Docking of a homology model of the upstream acyl carrier protein (ACP6) against the TE suggests that the 2-fold axis of the TE dimer may also be the axis of symmetry that determines the arrangement of domains in the entire DEBS. Sequence conservation suggests that all TEs from modular polyketide synthases have a similar fold, dimer 2-fold axis, and substrate channel geometry.
- Department of Biochemistry and Biophysics, University of California, San Francisco, CA 94143, USA.
Organizational Affiliation: