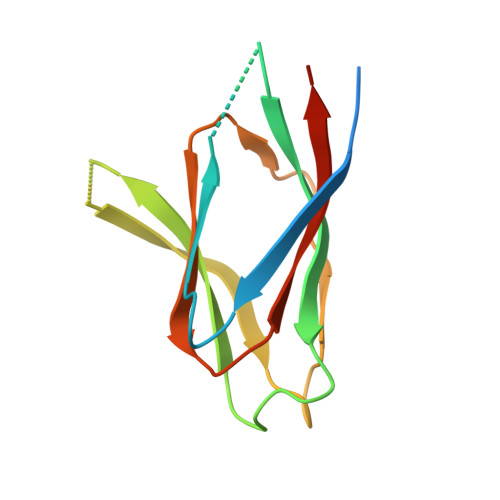
The crystal structure of nucleoplasmin-core: implications for histone binding and nucleosome assembly.
Dutta, S., Akey, I.V., Dingwall, C., Hartman, K.L., Laue, T., Nolte, R.T., Head, J.F., Akey, C.W.(2001) Mol Cell 8: 841-853
- PubMed: 11684019
- DOI: https://doi.org/10.1016/s1097-2765(01)00354-9
- Primary Citation of Related Structures:
1K5J - PubMed Abstract:
The efficient assembly of histone complexes and nucleosomes requires the participation of molecular chaperones. Currently, there is a paucity of data on their mechanism of action. We now present the structure of an N-terminal domain of nucleoplasmin (Np-core) at 2.3 A resolution. The Np-core monomer is an eight-stranded beta barrel that fits snugly within a stable pentamer. In the crystal, two pentamers associate to form a decamer. We show that both Np and Np-core are competent to assemble large complexes that contain the four core histones. Further experiments and modeling suggest that these complexes each contain five histone octamers which dock to a central Np decamer. This work has important ramifications for models of histone storage, sperm chromatin decondensation, and nucleosome assembly.
- Department of Physiology and Biophysics, Boston University School of Medicine, 700 Albany Street, Boston, MA 02118, USA.
Organizational Affiliation: