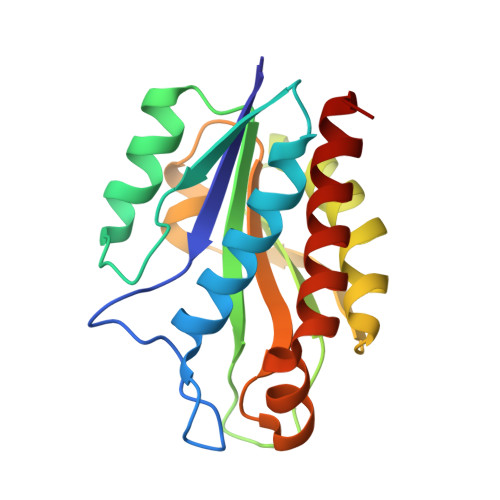
The Structure of the catalytic domain of N-acetylmuramoyl-L-alanine amidase, a cell wall hydrolase from Bacillus polymyxa var.colistinus and its resemblance to the structure of carboxypeptidases
Yamane, T., Koyama, Y., Nojiri, Y., Hikage, T., Akita, M., Suzuki, A., Shirai, T., Ise, F., Shida, T., Sekiguchi, J.To be published.