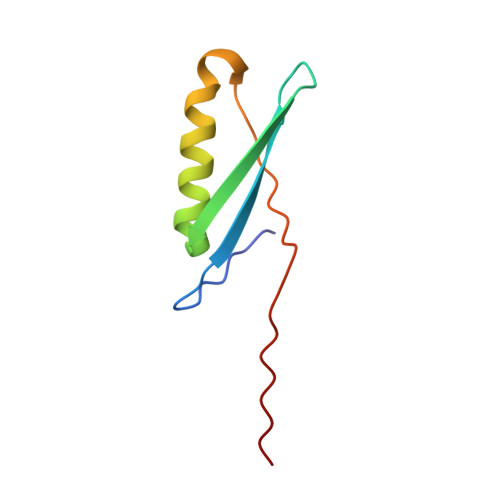
Conversion of monomeric protein L to an obligate dimer by computational protein design.
Kuhlman, B., O'Neill, J.W., Kim, D.E., Zhang, K.Y., Baker, D.(2001) Proc Natl Acad Sci U S A 98: 10687-10691
- PubMed: 11526208
- DOI: https://doi.org/10.1073/pnas.181354398
- Primary Citation of Related Structures:
1JML - PubMed Abstract:
Protein L consists of a single alpha-helix packed on a four-stranded beta-sheet formed by two symmetrically opposed beta-hairpins. We use a computer-based protein design procedure to stabilize a domain-swapped dimer of protein L in which the second beta-turn straightens and the C-terminal strand inserts into the beta-sheet of the partner. The designed obligate dimer contains three mutations (A52V, N53P, and G55A) and has a dissociation constant of approximately 700 pM, which is comparable to the dissociation constant of many naturally occurring protein dimers. The structure of the dimer has been determined by x-ray crystallography and is close to the in silico model.
- Department of Biochemistry and Howard Hughes Medical Institute, University of Washington, Seattle, WA 98195, USA.
Organizational Affiliation: