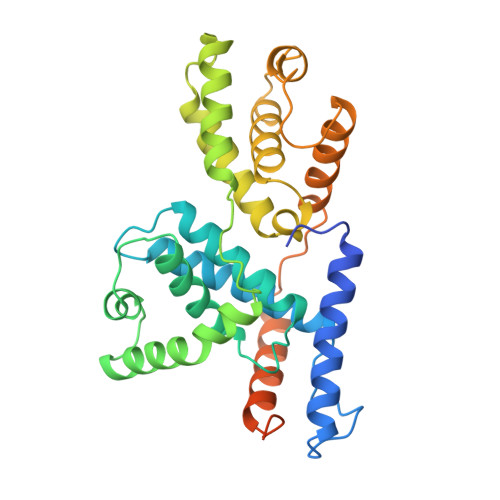
The crystal structure of human cyclin H.
Andersen, G., Poterszman, A., Egly, J.M., Moras, D., Thierry, J.C.(1996) FEBS Lett 397: 65-69
- PubMed: 8941715
- DOI: https://doi.org/10.1016/s0014-5793(96)01143-x
- Primary Citation of Related Structures:
1JKW - PubMed Abstract:
The crystal structure of human cyclin H has been solved at 2.6 A resolution by the MIR method and refined to an R-factor of 23.1%. The core of the molecule consists of two helical repeats adopting the canonical cyclin fold already observed in the structures of cyclin A [Brown et al. (1995) Structure 3, 1235-1247; Jeffrey et al. (1995) Nature 376, 313-320; Russo et al. (1996) Nature 382, 325-331] and TFIIB [Nikoilov et al. (1995) Nature 377, 119-128]. The N-terminal and C-terminal residues form a new domain built on two long helices interacting essentially with the first repeat of the molecule.
- Institut de Génétique et Biologie Moléculaire et Cellulaire, CNRS/INSERM/ULP, Illkirch, France.
Organizational Affiliation: