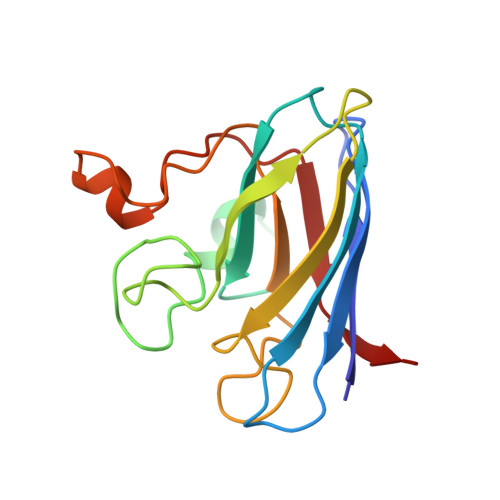
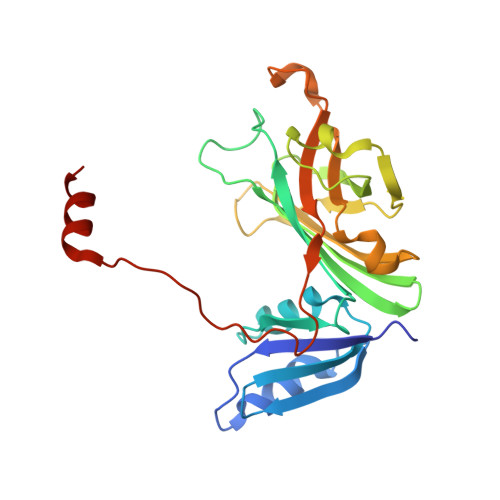
Heterodimeric structure of superoxide dismutase in complex with its metallochaperone.
Lamb, A.L., Torres, A.S., O'Halloran, T.V., Rosenzweig, A.C.(2001) Nat Struct Biol 8: 751-755
- PubMed: 11524675
- DOI: https://doi.org/10.1038/nsb0901-751
- Primary Citation of Related Structures:
1JK9 - PubMed Abstract:
The copper chaperone for superoxide dismutase (CCS) activates the eukaryotic antioxidant enzyme copper, zinc superoxide dismutase (SOD1). The 2.9 A resolution structure of yeast SOD1 complexed with yeast CCS (yCCS) reveals that SOD1 interacts with its metallochaperone to form a complex comprising one monomer of each protein. The heterodimer interface is remarkably similar to the SOD1 and yCCS homodimer interfaces. Striking conformational rearrangements are observed in both the chaperone and target enzyme upon complex formation, and the functionally essential C-terminal domain of yCCS is well positioned to play a key role in the metal ion transfer mechanism. This domain is linked to SOD1 by an intermolecular disulfide bond that may facilitate or regulate copper delivery.
- Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, Illinois 60208, USA.
Organizational Affiliation: