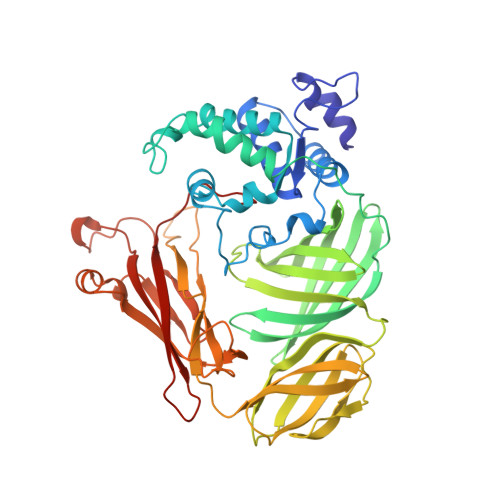
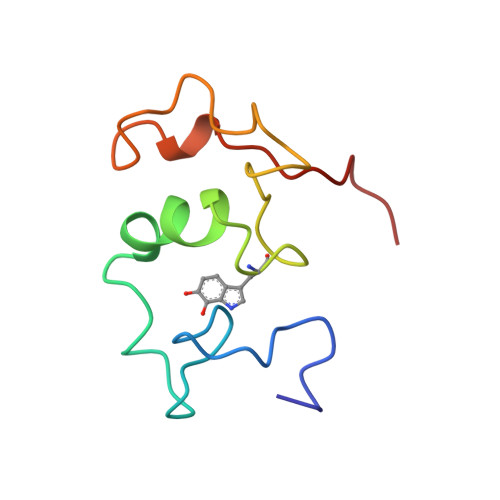
Structure of a quinohemoprotein amine dehydrogenase with an uncommon redox cofactor and highly unusual crosslinking.
Datta, S., Mori, Y., Takagi, K., Kawaguchi, K., Chen, Z.W., Okajima, T., Kuroda, S., Ikeda, T., Kano, K., Tanizawa, K., Mathews, F.S.(2001) Proc Natl Acad Sci U S A 98: 14268-14273
- PubMed: 11717396
- DOI: https://doi.org/10.1073/pnas.241429098
- Primary Citation of Related Structures:
1JJU - PubMed Abstract:
The crystal structure of the heterotrimeric quinohemoprotein amine dehydrogenase from Paracoccus denitrificans has been determined at 2.05-A resolution. Within an 82-residue subunit is contained an unusual redox cofactor, cysteine tryptophylquinone (CTQ), consisting of an orthoquinone-modified tryptophan side chain covalently linked to a nearby cysteine side chain. The subunit is surrounded on three sides by a 489-residue, four-domain subunit that includes a diheme cytochrome c. Both subunits sit on the surface of a third subunit, a 337-residue seven-bladed beta-propeller that forms part of the enzyme active site. The small catalytic subunit is internally crosslinked by three highly unusual covalent cysteine to aspartic or glutamic acid thioether linkages in addition to the cofactor crossbridge. The catalytic function of the enzyme as well as the biosynthesis of the unusual catalytic subunit is discussed.
- Department of Biochemistry and Molecular Biophysics, Washington University School of Medicine, St. Louis, MO 63110, USA.
Organizational Affiliation: