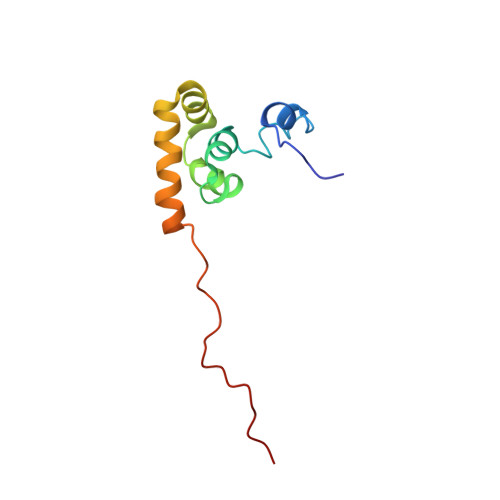
Structural basis of ligand recognition by PABC, a highly specific peptide-binding domain found in poly(A)-binding protein and a HECT ubiquitin ligase
Kozlov, G., De Crescenzo, G., Lim, N.S., Siddiqui, N., Fantus, D., Kahvejian, A., Trempe, J.F., Elias, D., Ekiel, I., Sonenberg, N., O'Connor-McCourt, M., Gehring, K.(2004) EMBO J 23: 272-281
- PubMed: 14685257
- DOI: https://doi.org/10.1038/sj.emboj.7600048
- Primary Citation of Related Structures:
1JGN, 1JH4 - PubMed Abstract:
The C-terminal domain of poly(A)-binding protein (PABC) is a peptide-binding domain found in poly(A)-binding proteins (PABPs) and a HECT (homologous to E6-AP C-terminus) family E3 ubiquitin ligase. In protein synthesis, the PABC domain of PABP functions to recruit several translation factors possessing the PABP-interacting motif 2 (PAM2) to the mRNA poly(A) tail. We have determined the solution structure of the human PABC domain in complex with two peptides from PABP-interacting protein-1 (Paip1) and Paip2. The structures show a novel mode of peptide recognition, in which the peptide binds as a pair of beta-turns with extensive hydrophobic, electrostatic and aromatic stacking interactions. Mutagenesis of PABC and peptide residues was used to identify key protein-peptide interactions and quantified by isothermal calorimetry, surface plasmon resonance and GST pull-down assays. The results provide insight into the specificity of PABC in mediating PABP-protein interactions.
- Department of Biochemistry, McGill University, 3655 Promenade Sir William Osler, Montreal, Quebec, Canada.
Organizational Affiliation: