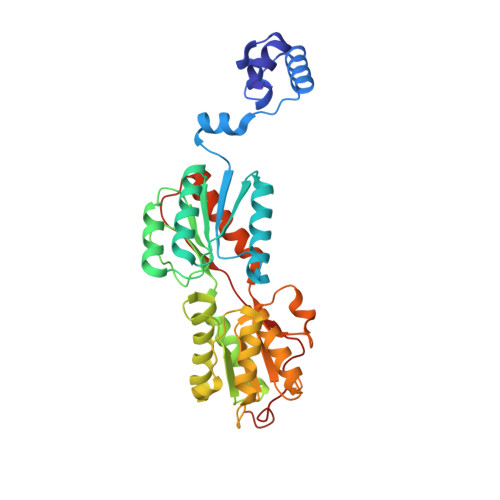
Role of residue 147 in the gene regulatory function of the Escherichia coli purine repressor.
Huffman, J.L., Lu, F., Zalkin, H., Brennan, R.G.(2002) Biochemistry 41: 511-520
- PubMed: 11781089
- DOI: https://doi.org/10.1021/bi0156660
- Primary Citation of Related Structures:
1JFS, 1JFT, 1JH9, 1JHZ - PubMed Abstract:
The crystal structures of corepressor-bound and free Escherichia coli purine repressor (PurR) have delineated the roles of several residues in corepressor binding and specificity and the intramolecular signal transduction (allosterism) of this LacI/GalR family member. From these structures, residue W147 was implicated as a key component of the allosteric response, but in many members of the LacI/GalR family, position 147 is occupied by an arginine. To understand the role of this tryptophan at position 147, three proteins, substituted by phenylalanine (W147F), alanine (W147A), or arginine (W147R), were constructed and characterized in vivo and in vitro, and their structures were determined. W147F displays a decreased affinity for corepressor and is a poor repressor in vivo. W147A and W147R, on the other hand, are super repressors and bind corepressor 13.6 and 7.9 times more tightly, respectively, than wild-type. Each mutant PurR-hypoxanthine-purF operator holo complex crystallizes isomorphously to wild-type. Whereas the apo corepressor binding domain (CBD) of W147F crystallizes under those conditions used for the wild-type protein, neither the apo CBD of W147R nor W147A crystallizes, although screened extensively for new crystal forms. Structures of the holo repressor mutants have been solved to resolutions between 2.5 and 2.9 A, and the structure of the apo CBD of W147F has been solved to 2.4 A resolution. These structures provide insight into the altered biochemical properties and physiological functions of these mutants, which appear to depend on the sometimes subtle preference for one conformation (apo vs holo) over the other.
- Department of Biochemistry and Molecular Biology, Oregon Health and Science University, Portland, Oregon, USA.
Organizational Affiliation: