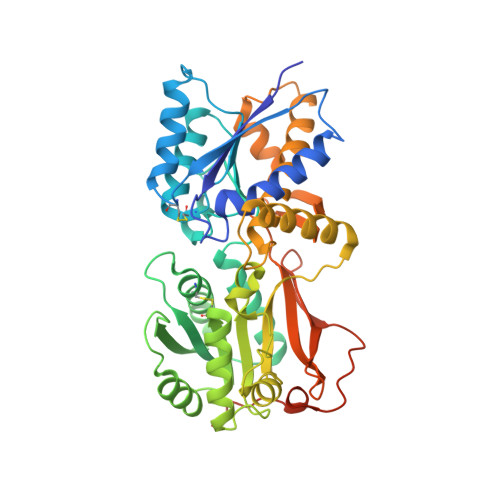
Allosteric activation of a spring-loaded natriuretic peptide receptor dimer by hormone.
He, X.l., Chow, D.c., Martick, M.M., Garcia, K.C.(2001) Science 293: 1657-1662
- PubMed: 11533490
- DOI: https://doi.org/10.1126/science.1062246
- Primary Citation of Related Structures:
1JDN, 1JDP - PubMed Abstract:
Natriuretic peptides (NPs) are vasoactive cyclic-peptide hormones important in blood pressure regulation through interaction with natriuretic cell-surface receptors. We report the hormone-binding thermodynamics and crystal structures at 2.9 and 2.0 angstroms, respectively, of the extracellular domain of the unliganded human NP receptor (NPR-C) and its complex with CNP, a 22-amino acid NP. A single CNP molecule is bound in the interface of an NPR-C dimer, resulting in asymmetric interactions between the hormone and the symmetrically related receptors. Hormone binding induces a 20 angstrom closure between the membrane-proximal domains of the dimer. In each monomer, the opening of an interdomain cleft, which is tethered together by a linker peptide acting as a molecular spring, is likely a conserved allosteric trigger for intracellular signaling by the natriuretic receptor family.
- Departments of Microbiology and Immunology and Structural Biology, Stanford University School of Medicine, Fairchild D319, 299 Campus Drive, Stanford, CA 93405-5124, USA.
Organizational Affiliation: