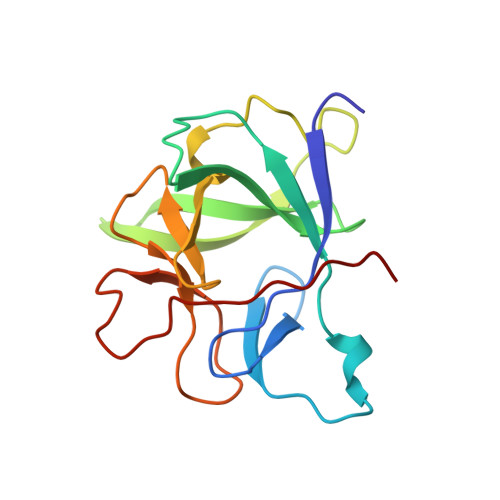
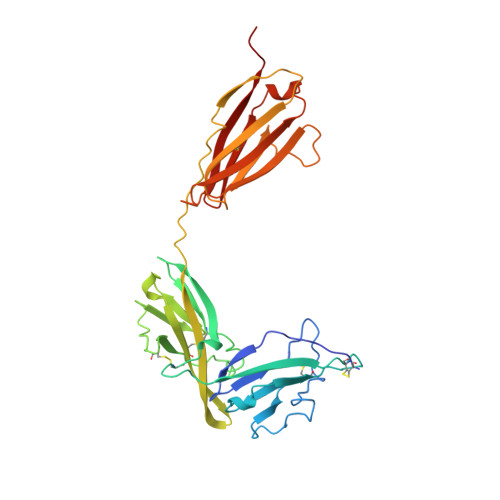
Crystal structure of the type-I interleukin-1 receptor complexed with interleukin-1beta.
Vigers, G.P., Anderson, L.J., Caffes, P., Brandhuber, B.J.(1997) Nature 386: 190-194
- PubMed: 9062193
- DOI: https://doi.org/10.1038/386190a0
- Primary Citation of Related Structures:
1ITB - PubMed Abstract:
Interleukin-1 (IL-1) is an important mediator of inflammatory disease. The IL-1 family currently consists of two agonists, IL-1alpha and IL-1beta, and one antagonist, IL-1ra. Each of these molecules binds to the type I IL-1 receptor (IL1R). The binding of IL-1alpha or IL-1beta to IL1R is an early step in IL-1 signal transduction and blocking this interaction may therefore be a useful target for the development of new drugs. Here we report the three-dimensional structure of IL-1beta bound to the extracellular domain of IL1R (s-IL1R) at 2.5 A resolution. IL-1beta binds to s-IL1R with a 1:1 stoichiometry. The crystal structure shows that s-IL1R consists of three immunoglobulin-like domains which wrap around IL-1beta in a manner distinct from the structures of previously described cytokine-receptor complexes. The two receptor-binding regions on IL-1beta identified by site-directed mutagenesis both contact the receptor: one binds to the first two domains of the receptor, while the other binds exclusively to the third domain.
- Amgen Inc., Boulder, Colorado 80301, USA.
Organizational Affiliation: