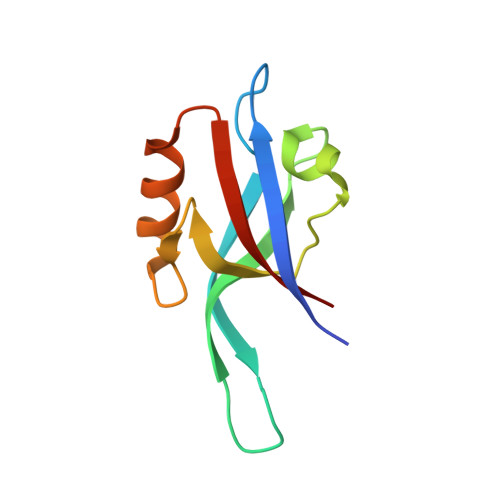
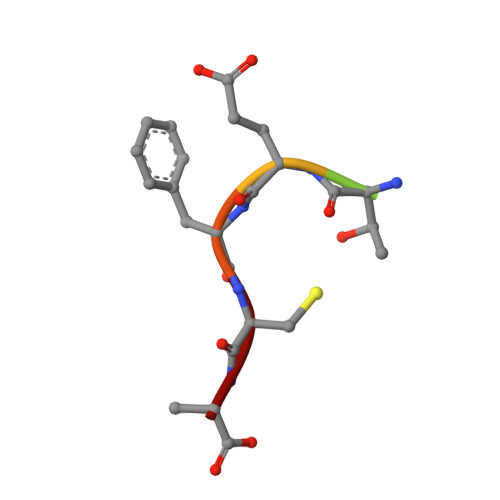
Functional relevance of the disulfide-linked complex of the N-terminal PDZ domain of InaD with NorpA.
Kimple, M.E., Siderovski, D.P., Sondek, J.(2001) EMBO J 20: 4414-4422
- PubMed: 11500369
- DOI: https://doi.org/10.1093/emboj/20.16.4414
- Primary Citation of Related Structures:
1IHJ - PubMed Abstract:
In Drosophila, phototransduction is mediated by G(q)-activation of phospholipase C and is a well studied model system for understanding the kinetics of signal initiation, propagation and termination controlled by G proteins. The proper intracellular targeting and spatial arrangement of most proteins involved in fly phototransduction require the multi-domain scaffolding protein InaD, composed almost entirely of five PDZ domains, which independently bind various proteins including NorpA, the relevant phospho lipase C-beta isozyme. We have determined the crystal structure of the N-terminal PDZ domain of InaD bound to a peptide corresponding to the C-terminus of NorpA to 1.8 A resolution. The structure highlights an intermolecular disulfide bond necessary for high affinity interaction as determined by both in vitro and in vivo studies. Since other proteins also possess similar, cysteine-containing consensus sequences for binding PDZ domains, this disulfide-mediated 'dock-and-lock' interaction of PDZ domains with their ligands may be a relatively ubiquitous mode of coordinating signaling pathways.
- Department of Biochemistry and Biophysics, The University of North Carolina at Chapel Hill, Chapel Hill, NC 27599, USA.
Organizational Affiliation: