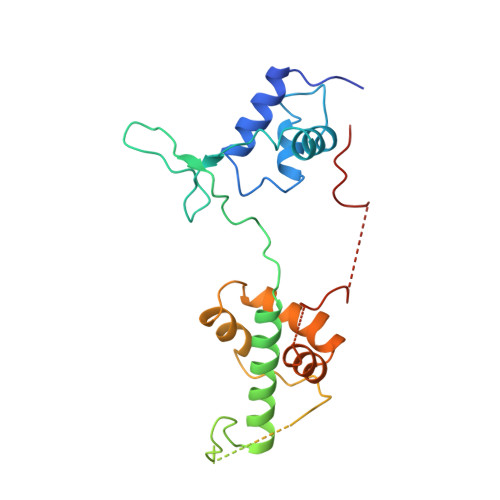
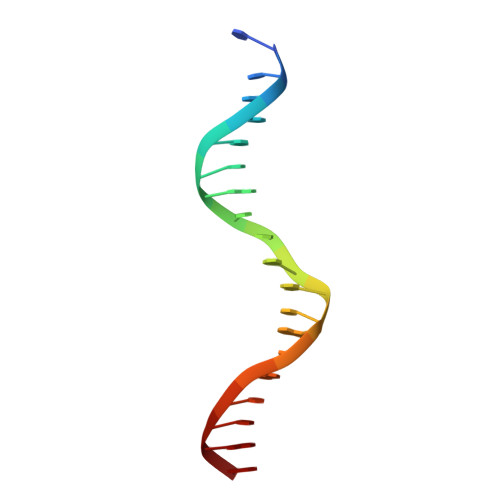
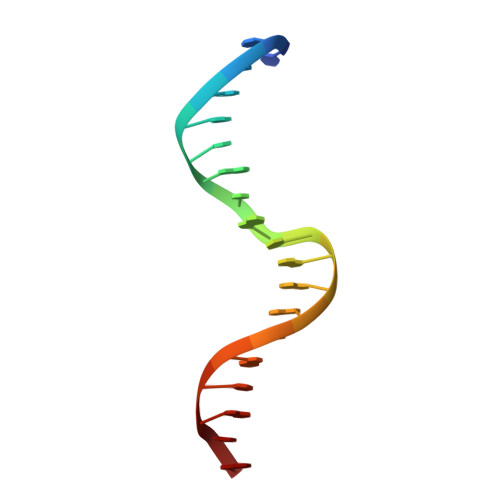
The crystal structure of the DNA-binding domain of yeast RAP1 in complex with telomeric DNA.
Konig, P., Giraldo, R., Chapman, L., Rhodes, D.(1996) Cell 85: 125-136
- PubMed: 8620531
- DOI: https://doi.org/10.1016/s0092-8674(00)81088-0
- Primary Citation of Related Structures:
1IGN - PubMed Abstract:
Telomeres, the nucleoprotein complexes at the ends of eukaryotic chromosomes, are essential for chromosome stability. In the yeast S. cerevisiae, telomeric DNA is bound in a sequence-specific manner by RAP1, a multifunctional protein also involved in transcriptional regulation. Here we report the crystal structure of the DNA-binding domain of RAP1 in complex with telomeric DNA site at 2.25 A resolution. The protein contains two similar domains that bind DNA in a tandem orientation, recognizing a tandemly repeated DNA sequence. The domains are structurally related to the homeodomain and the proto-oncogene Myb, but show novel features in their DNA-binding mode. A structured linker between the domains and a long C-terminal tail contribute to the binding specificity. This structure provides insight into the recognition of the conserved telomeric DNA sequences by a protein.
- Medical Research Council, Laboratory of Molecular Biology, Cambridge, United Kingdom.
Organizational Affiliation: