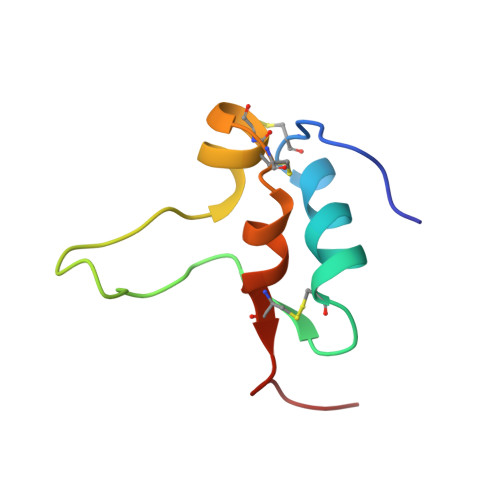
Solution structure of human insulin-like growth factor II. Relationship to receptor and binding protein interactions.
Torres, A.M., Forbes, B.E., Aplin, S.E., Wallace, J.C., Francis, G.L., Norton, R.S.(1995) J Mol Biology 248: 385-401
- PubMed: 7739048
- DOI: https://doi.org/10.1016/s0022-2836(95)80058-1
- Primary Citation of Related Structures:
1IGL - PubMed Abstract:
The three-dimensional structure of human insulin-like growth factor (IGF) II in aqueous solution at pH 3.1 and 300 K has been determined from nuclear magnetic resonance data and restrained molecular dynamics calculations. Structural constraints consisting of 502 NOE-derived distance constraints, 11 dihedral angle restraints, and three disulfide bridges were used as input for distance geometry calculations in DIANA and X-PLOR, followed by simulated annealing refinement and energy minimization in X-PLOR. The resulting family of 20 structures was well defined in the regions of residues 5 to 28 and 41 to 62, with an average pairwise root-mean-square deviation of 1.24 A for the backbone heavy-atoms (N, C2, C) and 1.90 A for all heavy atoms. The poorly defined regions consist of the N and C termini, part of the B-domain, and the C-domain loop. Resonances from these regions of the protein gave stronger cross peaks in two dimensional NMR spectra, consistent with significant motional averaging. The main secondary structure elements in IGF-II are alpha-helices encompassing residues 11 to 21, 42 to 49 and 53 to 59. A small anti-parallel beta-sheet is formed by residues 59 to 61 and 25 to 27, while residues 26 to 28 appear to participate in intermolecular beta-sheet formation. The structure of IGF-II in the well-defined regions is very similar to those of the corresponding regions of insulin and IGF-I. Significant differences between IGF-II and IGF-I occur near the start of the third helix, in a region known to modulate affinity for the type 2 IGF receptor, and at the C terminus. The IGF II structure is discussed in relation to its binding sites for the insulin and IGF receptors and the IGF binding proteins.
- NMR Laboratory Biomolecular Research Institute, Parkville, Australia.
Organizational Affiliation: