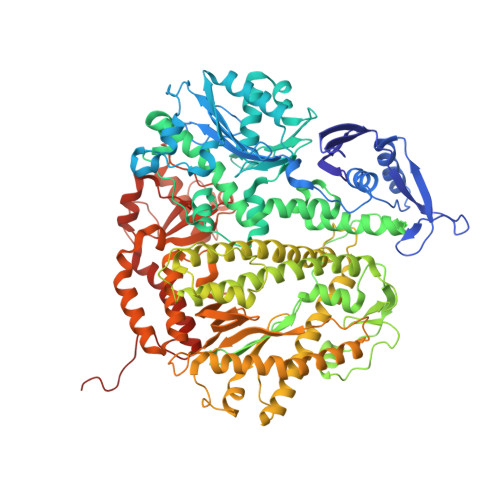
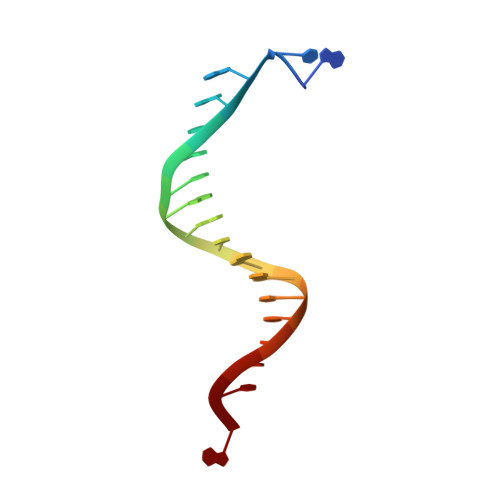
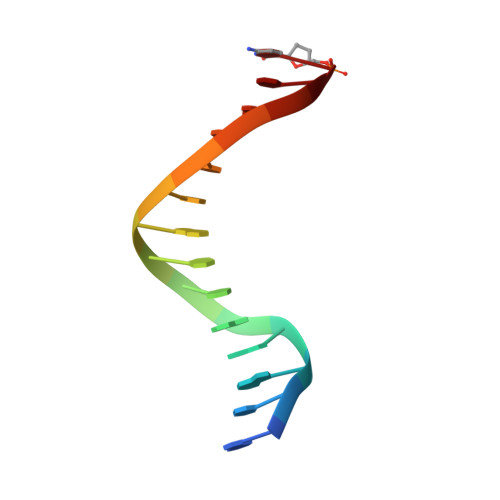
Structure of the Replicating Complex of a Pol Alpha Family DNA Polymerase
Franklin, M.C., Wang, J., Steitz, T.A.(2001) Cell 105: 657-667
- PubMed: 11389835
- DOI: https://doi.org/10.1016/s0092-8674(01)00367-1
- Primary Citation of Related Structures:
1IG9, 1IH7 - PubMed Abstract:
We describe the 2.6 A resolution crystal structure of RB69 DNA polymerase with primer-template DNA and dTTP, capturing the step just before primer extension. This ternary complex structure in the human DNA polymerase alpha family shows a 60 degrees rotation of the fingers domain relative to the apo-protein structure, similar to the fingers movement in pol I family polymerases. Minor groove interactions near the primer 3' terminus suggest a common fidelity mechanism for pol I and pol alpha family polymerases. The duplex product DNA orientation differs by 40 degrees between the polymerizing mode and editing mode structures. The role of the thumb in this DNA motion provides a model for editing in the pol alpha family.
- Department of Molecular Biophysics and Biochemistry, Yale University, New Haven, CT 06511, USA.
Organizational Affiliation: