A combinatorial approach for determining protease specificities: application to interleukin-1beta converting enzyme (ICE).
Rano, T.A., Timkey, T., Peterson, E.P., Rotonda, J., Nicholson, D.W., Becker, J.W., Chapman, K.T., Thornberry, N.A.(1997) Chem Biol 4: 149-155
- PubMed: 9190289
- DOI: https://doi.org/10.1016/s1074-5521(97)90258-1
- Primary Citation of Related Structures:
1IBC - PubMed Abstract:
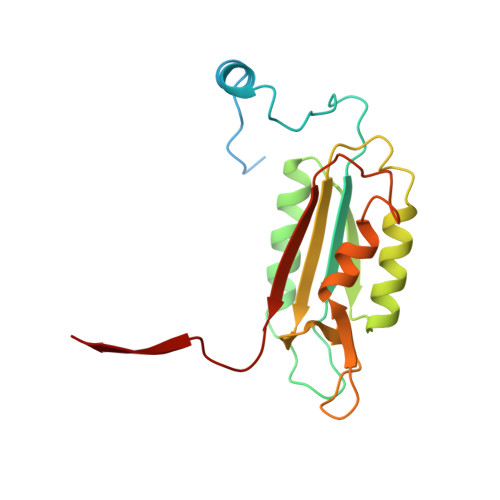
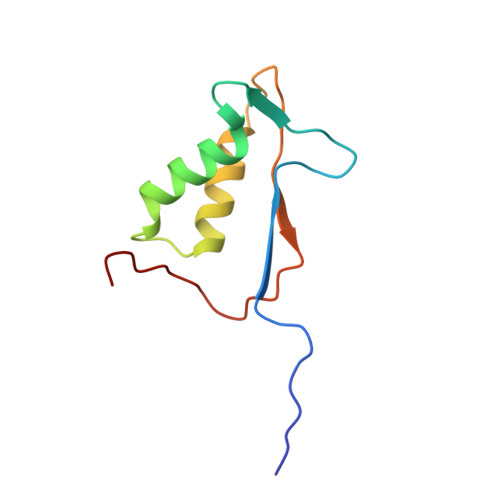
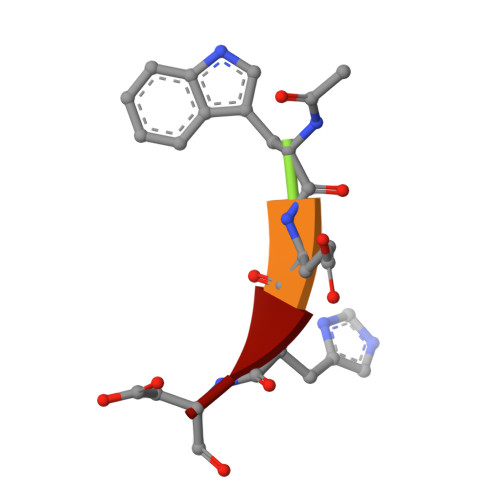
Interleukin-1beta converting enzyme (ICE/caspase-1) is the protease responsible for interleukin-1beta (IL-1beta) production in monocytes. It was the first member of a new cysteine protease family to be identified. Members of this family have functions in both inflammation and apoptosis. A novel method for identifying protease specificity, employing a positional-scanning substrate library, was used to determine the amino-acid preferences of ICE. Using this method, the complete specificity of a protease can be mapped in the time required to perform one assay. The results indicate that the optimal tetrapeptide recognition sequence for ICE is WEHD, not YVAD, as previously believed, and this led to the synthesis of an unusually potent aldehyde inhibitor, Ac-WEHD-CHO (Ki = 56 pM). The structural basis for this potent inhibition was determined by X-ray crystallography. The results presented in this study establish a positional-scanning library as a powerful tool for rapidly and accurately assessing protease specificity. The preferred sequence for ICE (WEHD) differs significantly from that found in human pro-interleukin-1beta (YVHD), which suggests that this protease may have additional endogenous substrates, consistent with evidence linking it to apoptosis and IL-1alpha production.
- Department of Molecular Design and Diversity, Merck Research Laboratories, R123-232, PO Box 2000, Rahway, New Jersey 07065, USA.
Organizational Affiliation: