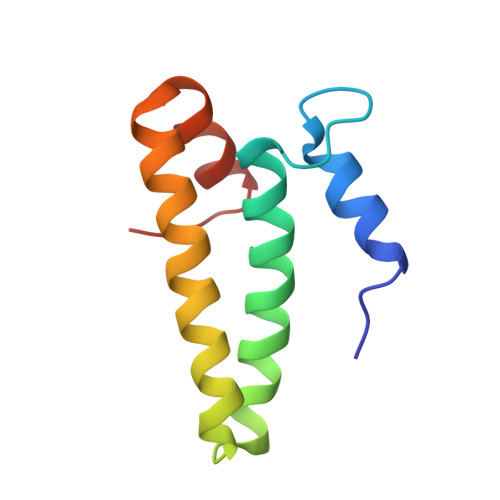
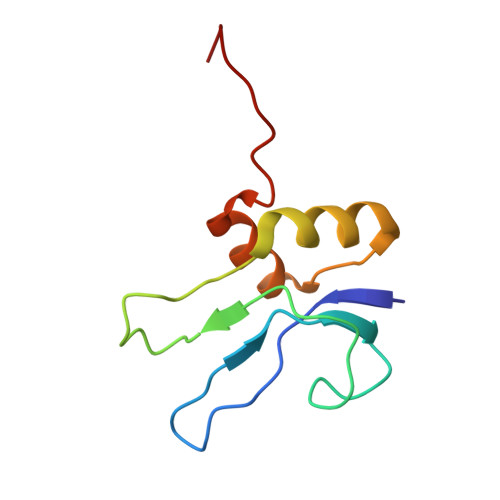
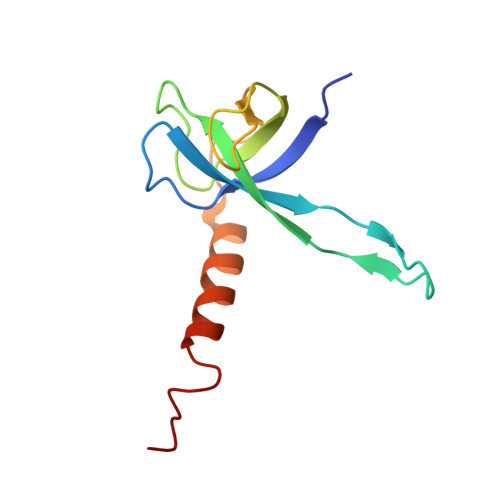
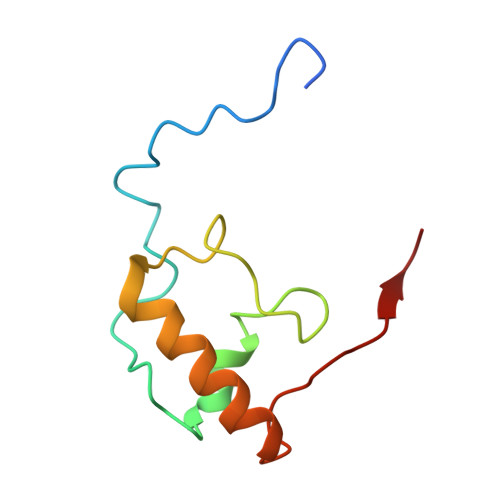
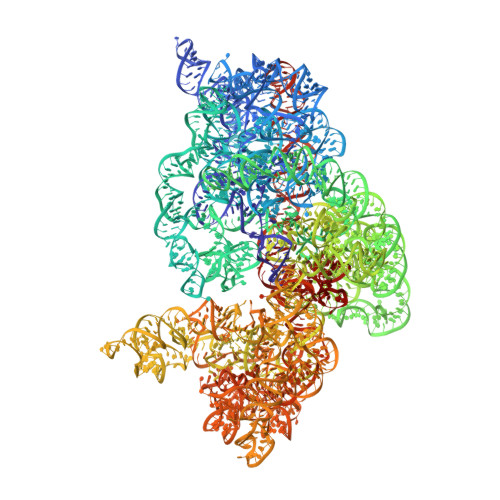
Crystal structures of complexes of the small ribosomal subunit with tetracycline, edeine and IF3.
Pioletti, M., Schlunzen, F., Harms, J., Zarivach, R., Gluhmann, M., Avila, H., Bashan, A., Bartels, H., Auerbach, T., Jacobi, C., Hartsch, T., Yonath, A., Franceschi, F.(2001) EMBO J 20: 1829-1839
- PubMed: 11296217
- DOI: https://doi.org/10.1093/emboj/20.8.1829
- Primary Citation of Related Structures:
1I94, 1I95, 1I96, 1I97 - PubMed Abstract:
The small ribosomal subunit is responsible for the decoding of genetic information and plays a key role in the initiation of protein synthesis. We analyzed by X-ray crystallography the structures of three different complexes of the small ribosomal subunit of Thermus thermophilus with the A-site inhibitor tetracycline, the universal initiation inhibitor edeine and the C-terminal domain of the translation initiation factor IF3. The crystal structure analysis of the complex with tetracycline revealed the functionally important site responsible for the blockage of the A-site. Five additional tetracycline sites resolve most of the controversial biochemical data on the location of tetracycline. The interaction of edeine with the small subunit indicates its role in inhibiting initiation and shows its involvement with P-site tRNA. The location of the C-terminal domain of IF3, at the solvent side of the platform, sheds light on the formation of the initiation complex, and implies that the anti-association activity of IF3 is due to its influence on the conformational dynamics of the small ribosomal subunit.
- Max-Planck-Institut für Molekulare Genetik, Ihnestrasse 73, 14195 Berlin, Germany.
Organizational Affiliation: