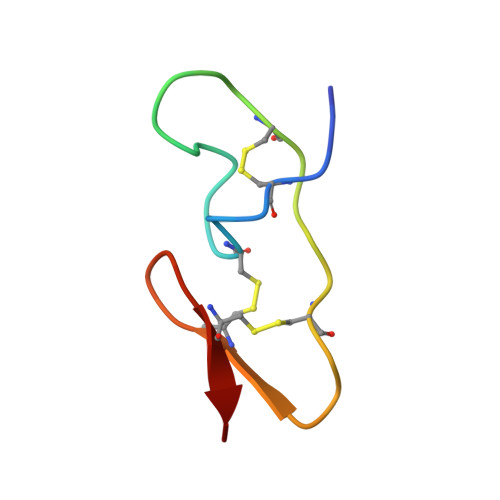
The structure of spider toxin huwentoxin-II with unique disulfide linkage: evidence for structural evolution.
Shu, Q., Lu, S.Y., Gu, X.C., Liang, S.P.(2002) Protein Sci 11: 245-252
- PubMed: 11790834
- DOI: https://doi.org/10.1110/ps.30502
- Primary Citation of Related Structures:
1I25 - PubMed Abstract:
The three-dimensional structure of huwentoxin-II (HWTX-II), an insecticidal peptide purified from the venom of spider Selenocosmia huwena with a unique disulfide bond linkage as I-III, II-V, and IV-VI, has been determined using 2D (1)H-NMR. The resulting structure of HWTX-II contains two beta-turns (C4-S7 and K24-W27) and a double-stranded antiparallel beta-sheet (W27-C29 and C34-K36). Although the C-terminal double-stranded beta-sheet cross-linked by two disulfide bonds (II-V and IV-VI in HWTX-II, II-V and III-VI in the ICK molecules) is conserved both in HWTX-II and the ICK molecules, the structure of HWTX-II is unexpected absence of the cystine knot because of its unique disulfide linkage. It suggests that HWTX-II adopts a novel scaffold different from the ICK motif that is adopted by all other spider toxin structures elucidated thus far. Furthermore, the structure of HWTX-II, which conforms to the disulfide-directed beta-hairpin (DDH) motif, not only supports the hypothesis that the ICK is a minor elaboration of the more ancestral DDH motif but also suggests that HWTX-II may have evolved from the same structural ancestor.
- Life Science College, Peking University, Beijing 100871, People's Republic of China.
Organizational Affiliation: