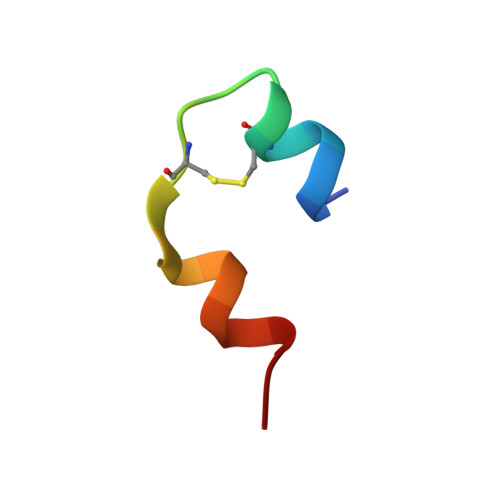
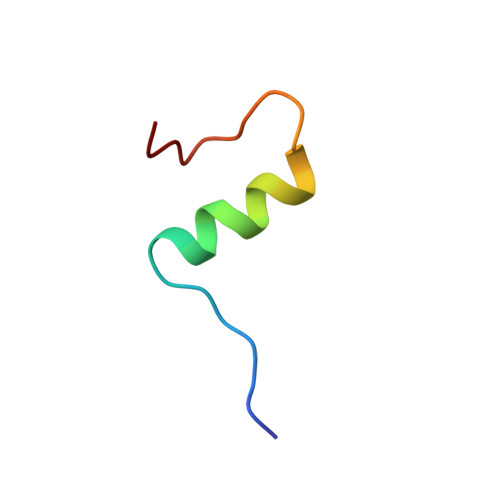
Solution structure of an engineered insulin monomer at neutral pH.
Olsen, H.B., Ludvigsen, S., Kaarsholm, N.C.(1996) Biochemistry 35: 8836-8845
- PubMed: 8688419
- DOI: https://doi.org/10.1021/bi960292+
- Primary Citation of Related Structures:
1HUI - PubMed Abstract:
Insulin circulates in the bloodstream and binds to its specific cell-surface receptor as a 5808 Da monomeric species. However, studies of the monomer structure and dynamics in solution are severely limited by insulin self-association into dimers and higher oligomers. In the present work we use site-directed mutagenesis of the dimer- and hexamer-forming surfaces to yield the first insulin species amenable for structure determination at neutral pH by nuclear magnetic resonance (NMR) spectroscopy. The preferred insulin mutant, i.e., (B1, B10, B16, B27) Glu, des-B30 insulin retains 47% biological potency and remains monomeric at millimolar concentrations in aqueous solution at pH 6.5-7.5 as judged by NMR and near-UV circular dichroism (CD) spectroscopy. From a series of 2D 1H-NMR spectra collected at pH 6.5 and 34 degrees C, the majority of the resonances are assigned to specific residues in the sequence, and nuclear Overhauser enhancement (NOE) cross-peaks are identified. NOE-derived distance restraints in conjunction with torsion restraints based on measured coupling constants, 3JHNH alpha, are used for structure calculations using the hybrid method of distance geometry and simulated annealing. The calculated structures show that the major part of the insulin mutant is structurally well defined with an average root mean square (rms) deviation between the 25 calculated structures and the mean coordinates of 0.66 A for backbone atoms (A2-A19 and B4-B26) and 1.31 A for all backbone atoms. The A-chain consists of two antiparallel helices, A2-A7 and A12-A19, connected by a loop. The B-chain contains a loop region (B1-B8), an alpha-helix (B9-B19), and a type I turn (B20-B23) and terminates as an extended strand (B24-B29). The B1-B4 and B27-B29 regions are disordered in solution. The structure is generally similar to crystal structures and resembles a crystalline T-state more than an R-state in the sense that the B-chain helix is confined to residues B9-B19.
- Novo Research Institute, Novo Nordisk A/S, Bagsvaerd, Denmark.
Organizational Affiliation: