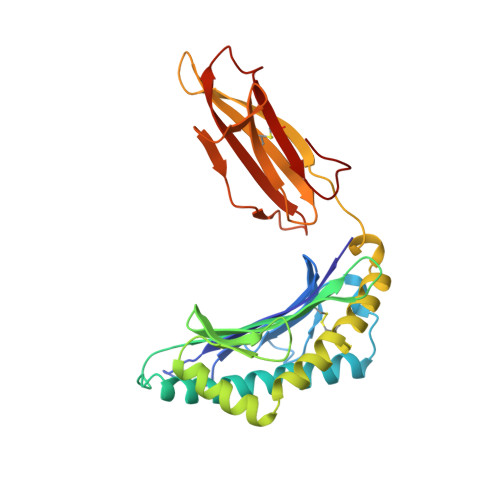
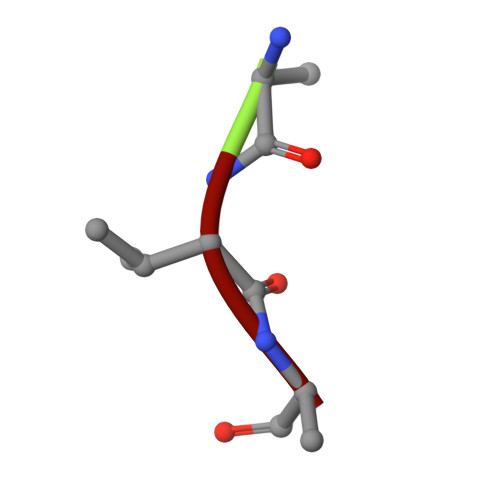
Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle.
Guo, H.C., Jardetzky, T.S., Garrett, T.P., Lane, W.S., Strominger, J.L., Wiley, D.C.(1992) Nature 360: 364-366
- PubMed: 1448153
- DOI: https://doi.org/10.1038/360364a0
- Primary Citation of Related Structures:
1HSB - PubMed Abstract:
We report here the determination and refinement to 1.9 A resolution by X-ray cryo-crystallography the structure of HLA-Aw68. The averaged image from the collection of bound, endogenous peptides clearly shows the atomic structure at the first three and last two amino acids in the peptides but no connected electron density in between. This suggests that bound peptides, held at both ends, take alternative pathways and could be of different lengths by bulging out in the middle. Peptides eluted from HLA-Aw68 include peptides of 9, 10 and 11 amino acids, a direct indication of the length heterogeneity of tightly bound peptides. Peptide sequencing shows relatively conserved 'anchor' residues at position 2 and the carboxy-terminal residue. Conserved binding sites for the peptide N and C termini at the ends of the class I major histocompatibility complex binding groove are apparently dominant in producing the long half-lives of peptide binding and the peptide-dependent stabilization of the class I molecule's structure.
- Department of Biochemistry and Molecular Biology, Harvard University, Cambridge, Massachusetts 02138.
Organizational Affiliation: