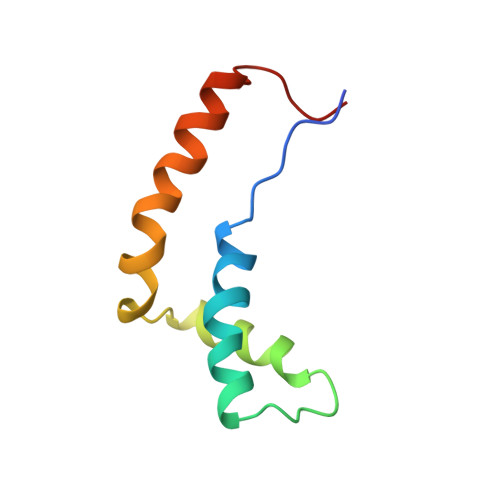
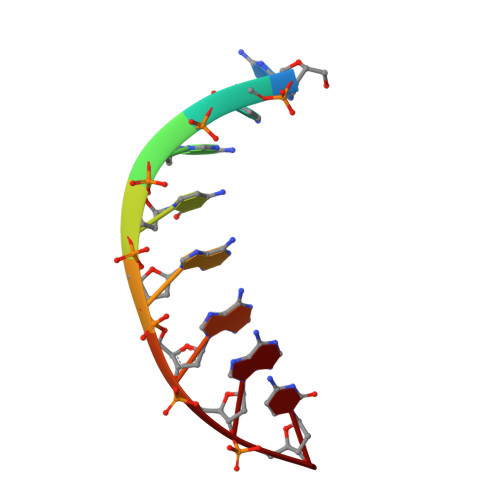
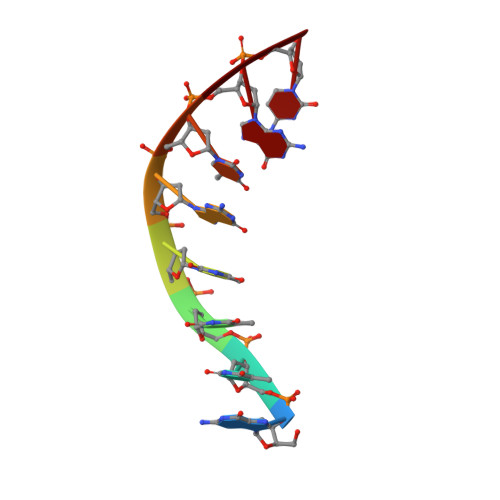
Molecular basis of human 46X,Y sex reversal revealed from the three-dimensional solution structure of the human SRY-DNA complex.
Werner, M.H., Huth, J.R., Gronenborn, A.M., Clore, G.M.(1995) Cell 81: 704-705
- PubMed: 7774012
- DOI: https://doi.org/10.1016/0092-8674(95)90532-4
- Primary Citation of Related Structures:
1HRY, 1HRZ - PubMed Abstract:
The solution structure of the specific complex between the high mobility group (HMG) domain of SRY (hSRY-HMG), the protein encoded by the human testis-determining gene, and its DNA target site in the promoter of the müllerian inhibitory substance gene has been determined by multidimensional NMR spectroscopy. hSRY-HMG has a twisted L shape that presents a concave surface (made up of three helices and the N- and C-terminal strands) to the DNA for sequence-specific recognition. Binding of hSRY-HMG to its specific target site occurs exclusively in the minor groove and induces a large conformational change in the DNA. The DNA in the complex has an overall 70 degrees-80 degrees bend and is helically unwound relative to classical A- and B-DNA. The structure of the complex reveals the origin of sequence-specific binding within the HMG-1/HMG-2 family and provides a framework for understanding the effects of point mutations that cause 46X,Y sex reversal at the atomic level.
- Laboratory of Chemical Physics, National Institute of Diabetes and Digestive and Kidney Diseases, National Institutes of Health, Bethesda, Maryland 20892-0520, USA.
Organizational Affiliation: