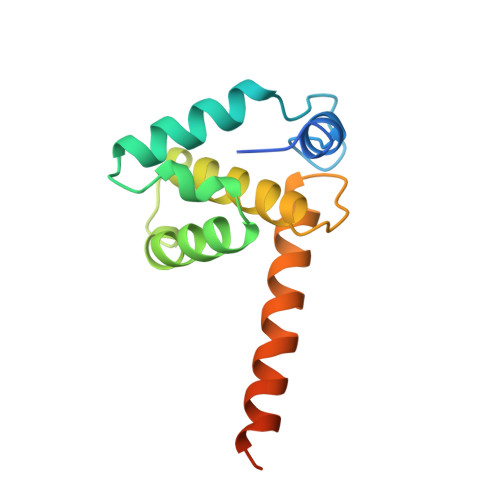
Crystal structures of the trimeric human immunodeficiency virus type 1 matrix protein: implications for membrane association and assembly.
Hill, C.P., Worthylake, D., Bancroft, D.P., Christensen, A.M., Sundquist, W.I.(1996) Proc Natl Acad Sci U S A 93: 3099-3104
- PubMed: 8610175
- DOI: https://doi.org/10.1073/pnas.93.7.3099
- Primary Citation of Related Structures:
1HIW - PubMed Abstract:
The human immunodeficiency virus type 1 (HIV-1) matrix protein forms a structural shell associated with the inner viral membrane and performs other essential functions throughout the viral life cycle. The crystal structure of the HIV-1 matrix protein, determined at 2.3 angstrom resolution, reveals that individual matrix molecules are composed of five major helices capped by a three-stranded mixed beta-sheet. Unexpectedly, the protein assembles into a trimer in three different crystal lattices, burying 1880 angstrom2 of accessible surface area at the trimer interfaces. Trimerization appears to create a large, bipartite membrane binding surface in which exposed basic residues could cooperate with the N-terminal myristoyl groups to anchor the protein on the acidic inner membrane of the virus.
- Department of Biochemistry, University of Utah, Salt Lake City, 84132, USA.
Organizational Affiliation: