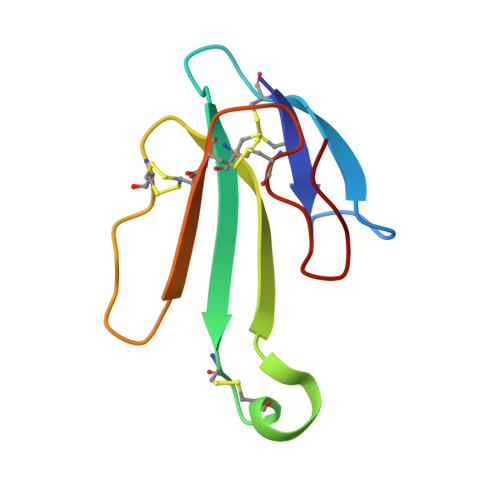
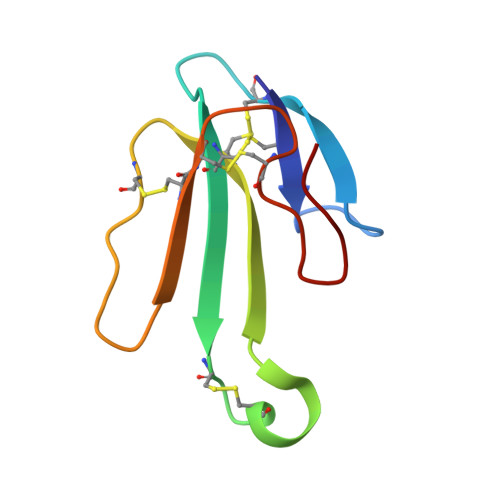
The Binding Site of Acetylcholine Receptor as Visualized in the X-Ray Structure of a Complex between Alpha-Bungarotoxin and a Mimotope Peptide.
Harel, M., Kasher, R., Nicolas, A., Guss, J.M., Balass, M., Fridkin, M., Smit, A.B., Brejc, K., Sixma, T.K., Katchalski-Katzir, E., Sussman, J.L., Fuchs, S.(2001) Neuron 32: 265
- PubMed: 11683996
- DOI: https://doi.org/10.1016/s0896-6273(01)00461-5
- Primary Citation of Related Structures:
1HC9 - PubMed Abstract:
We have determined the crystal structure at 1.8 A resolution of a complex of alpha-bungarotoxin with a high affinity 13-residue peptide that is homologous to the binding region of the alpha subunit of acetylcholine receptor. The peptide fits snugly to the toxin and adopts a beta hairpin conformation. The structures of the bound peptide and the homologous loop of acetylcholine binding protein, a soluble analog of the extracellular domain of acetylcholine receptor, are remarkably similar. Their superposition indicates that the toxin wraps around the receptor binding site loop, and in addition, binds tightly at the interface of two of the receptor subunits where it inserts a finger into the ligand binding site, thus blocking access to the acetylcholine binding site and explaining its strong antagonistic activity.
- Department of Structural Biology, Weizmann Institute of Science, 76100, Rehovot, Israel.
Organizational Affiliation: