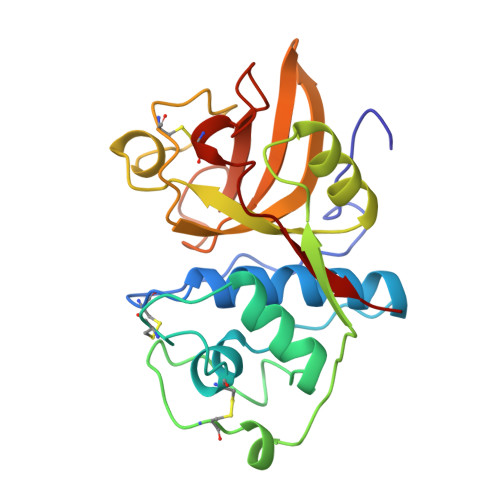
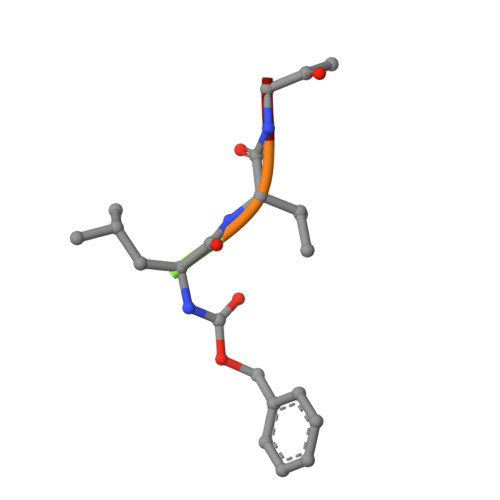
Crystal structure of glycyl endopeptidase from Carica papaya: a cysteine endopeptidase of unusual substrate specificity.
O'Hara, B.P., Hemmings, A.M., Buttle, D.J., Pearl, L.H.(1995) Biochemistry 34: 13190-13195
- PubMed: 7548082
- DOI: https://doi.org/10.1021/bi00040a034
- Primary Citation of Related Structures:
1GEC - PubMed Abstract:
Glycyl endopeptidase is a cysteine endopeptidase of the papain family, characterized by specificity for cleavage C-terminal to glycyl residues only and by resistance to inhibition by members of the cystatin family of cysteine proteinase inhibitors. Glycyl endopeptidase has been crystallized from high salt with a substrate-like inhibitor covalently bound to the catalytic Cys 25. The structure has been solved by molecular replacement with the structure of papain and refined at 2.1 A to an R factor of 0.196 (Rfree = 0.258) with good geometry. The structure of the S1 substrate binding site of glycyl endopeptidase differs from that of papain by the substitution of glycines at residues 23 and 65 in papain, with glutamic acid and arginine, respectively, in glycyl endopeptidase. The side chains of these residues form a barrier across the binding pocket, effectively excluding substrate residues with large side chains from the S1 subsite. The constriction of this subsite in glycyl endopeptidase explains the unique specificity of this enzyme for cleavage after glycyl residues and is a major component of its resistance to inhibition by cystatins.
- Department of Biochemistry and Molecular Biology, University College London, U.K.
Organizational Affiliation: