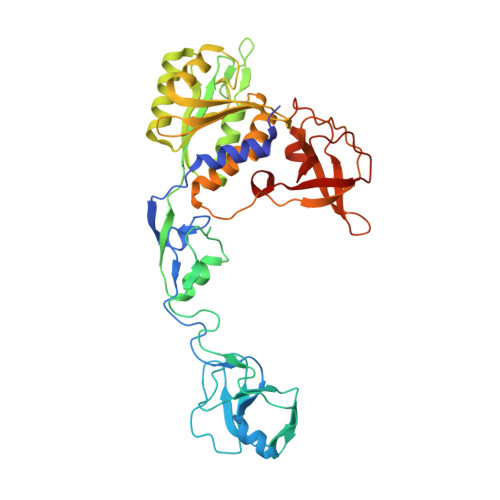
The crystal structure of Escherichia coli MoeA and its relationship to the multifunctional protein gephyrin.
Xiang, S., Nichols, J., Rajagopalan, K.V., Schindelin, H.(2001) Structure 9: 299-310
- PubMed: 11525167
- DOI: https://doi.org/10.1016/s0969-2126(01)00588-3
- Primary Citation of Related Structures:
1G8L, 1G8R - PubMed Abstract:
Molybdenum cofactor (Moco) biosynthesis is an evolutionarily conserved pathway present in archaea, eubacteria, and eukaryotes. In humans, genetic abnormalities in the biosynthetic pathway result in Moco deficiency, which is accompanied by severe neurological symptoms and death shortly after birth. The Escherichia coli MoeA and MogA proteins are involved in the final step of Moco biosynthesis: the incorporation of molybdenum into molybdopterin (MPT), the organic pyranopterin moiety of Moco. The crystal structure of E. coli MoeA has been refined at 2 A resolution and reveals that the highly elongated MoeA monomer consists of four clearly separated domains, one of which is structurally related to MogA, indicating a divergent evolutionary relationship between both proteins. The active form of MoeA is a dimer, and a putative active site appears to be localized to a cleft formed between domain II of the first monomer and domains III and IV of the second monomer. In eukaryotes, MogA and MoeA are fused into a single polypeptide chain. The corresponding mammalian protein gephyrin has also been implicated in the anchoring of glycinergic receptors to the cytoskeleton at inhibitory synapses. Based on the structures of MoeA and MogA, gephyrin is surmised to be a highly organized molecule containing at least five domains. This multidomain arrangement could provide a structural basis for its functional diversity. The oligomeric states of MoeA and MogA suggest how gephyrin could assemble into a hexagonal scaffold at inhibitory synapses.
- Department of Physiology and Biophysics, State University of New York at Stony Brook, 11794, USA.
Organizational Affiliation: