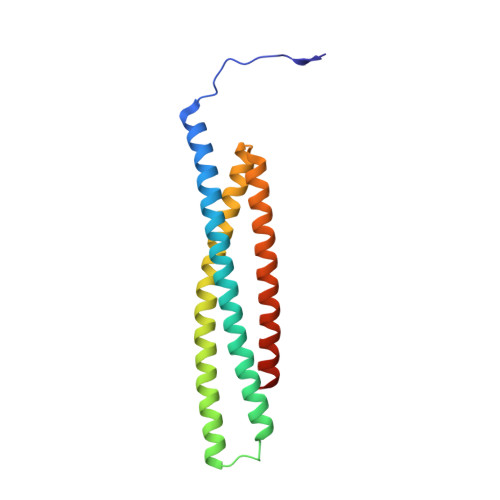
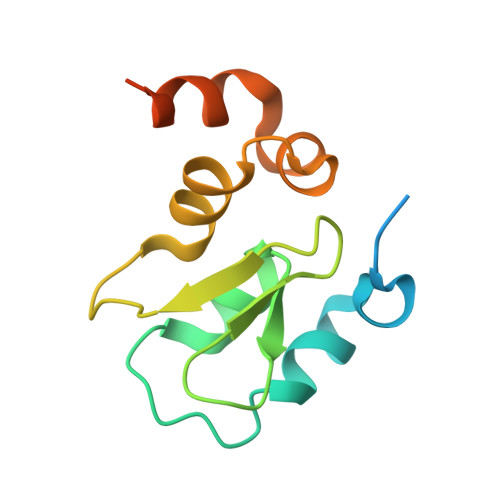
Structural basis of IAP recognition by Smac/DIABLO.
Wu, G., Chai, J., Suber, T.L., Wu, J.W., Du, C., Wang, X., Shi, Y.(2000) Nature 408: 1008-1012
- PubMed: 11140638
- DOI: https://doi.org/10.1038/35050012
- Primary Citation of Related Structures:
1G73 - PubMed Abstract:
Apoptosis is an essential process in the development and homeostasis of all metazoans. The inhibitor-of-apoptosis (IAP) proteins suppress cell death by inhibiting the activity of caspases; this inhibition is performed by the zinc-binding BIR domains of the IAP proteins. The mitochondrial protein Smac/DIABLO promotes apoptosis by eliminating the inhibitory effect of IAPs through physical interactions. Amino-terminal sequences in Smac/DIABLO are required for this function, as mutation of the very first amino acid leads to loss of interaction with IAPs and concomitant loss of Smac/DIABLO function. Here we report the high-resolution crystal structure of Smac/DIABLO complexed with the third BIR domain (BIR3) of XIAP. Our results show that the N-terminal four residues (Ala-Val-Pro-Ile) in Smac/DIABLO recognize a surface groove on BIR3, with the first residue Ala binding a hydrophobic pocket and making five hydrogen bonds to neighbouring residues on BIR3. These observations provide a structural explanation for the roles of the Smac N terminus as well as the conserved N-terminal sequences in the Drosophila proteins Hid/Grim/Reaper. In conjunction with other observations, our results reveal how Smac may relieve IAP inhibition of caspase-9 activity. In addition to explaining a number of biological observations, our structural analysis identifies potential targets for drug screening.
- Department of Molecular Biology, Princeton University, New Jersey 08544, USA.
Organizational Affiliation: