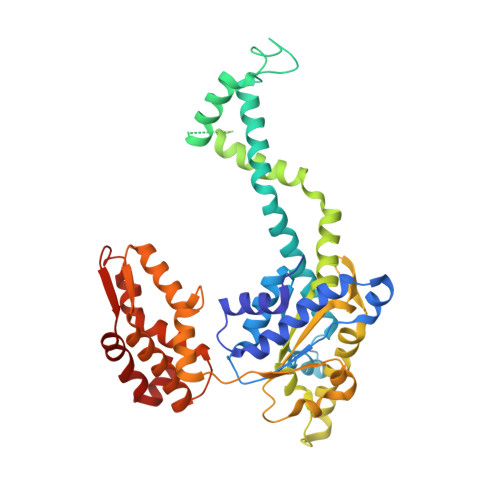
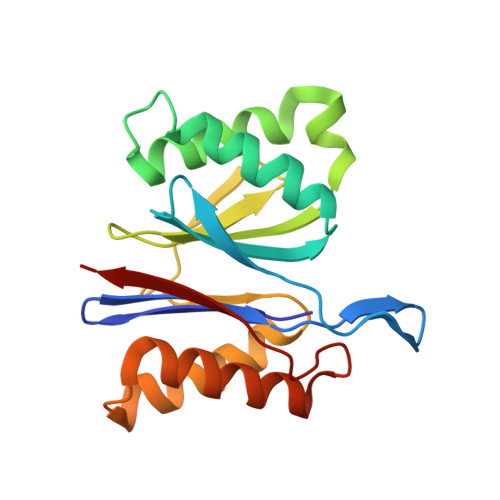
Crystal structures of the HslVU peptidase-ATPase complex reveal an ATP-dependent proteolysis mechanism.
Wang, J., Song, J.J., Franklin, M.C., Kamtekar, S., Im, Y.J., Rho, S.H., Seong, I.S., Lee, C.S., Chung, C.H., Eom, S.H.(2001) Structure 9: 177-184
- PubMed: 11250202
- DOI: https://doi.org/10.1016/s0969-2126(01)00570-6
- Primary Citation of Related Structures:
1G4A, 1G4B - PubMed Abstract:
The bacterial heat shock locus HslU ATPase and HslV peptidase together form an ATP-dependent HslVU protease. Bacterial HslVU is a homolog of the eukaryotic 26S proteasome. Crystallographic studies of HslVU should provide an understanding of ATP-dependent protein unfolding, translocation, and proteolysis by this and other ATP-dependent proteases. We present a 3.0 A resolution crystal structure of HslVU with an HslU hexamer bound at one end of an HslV dodecamer. The structure shows that the central pores of the ATPase and peptidase are next to each other and aligned. The central pore of HslU consists of a GYVG motif, which is conserved among protease-associated ATPases. The binding of one HslU hexamer to one end of an HslV dodecamer in the 3.0 A resolution structure opens both HslV central pores and induces asymmetric changes in HslV. Analysis of nucleotide binding induced conformational changes in the current and previous HslU structures suggests a protein unfolding-coupled translocation mechanism. In this mechanism, unfolded polypeptides are threaded through the aligned pores of the ATPase and peptidase and translocated into the peptidase central chamber.
- Department of Molecular Biophysics, Biochemistry, 266 Whitney Avenue, Yale University, 06520, New Haven, CT, USA. wang@mail.csb.yale.edu
Organizational Affiliation: