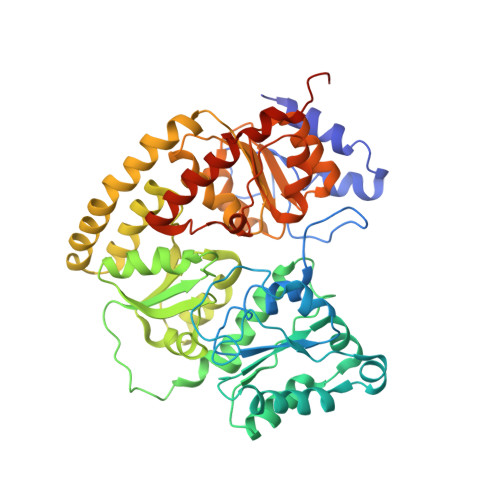
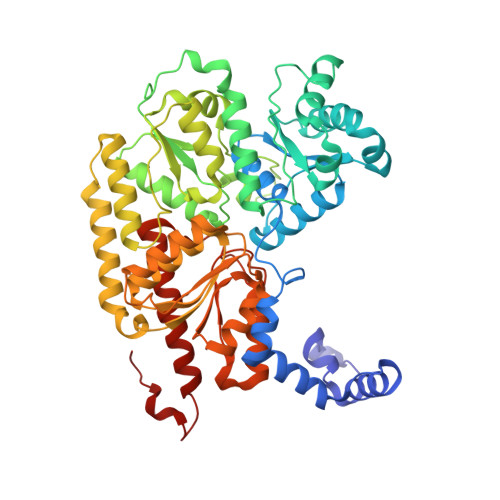
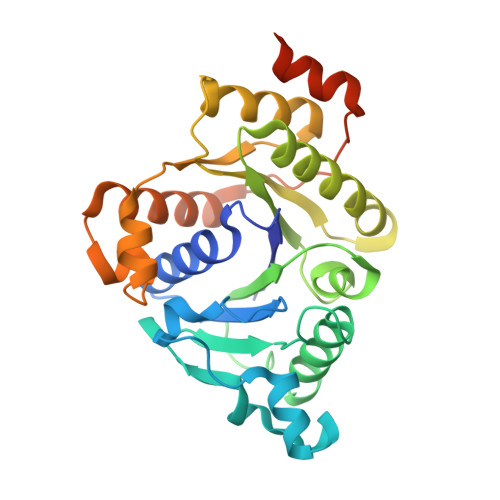
MgATP-Bound and nucleotide-free structures of a nitrogenase protein complex between the Leu 127 Delta-Fe-protein and the MoFe-protein.
Chiu, H., Peters, J.W., Lanzilotta, W.N., Ryle, M.J., Seefeldt, L.C., Howard, J.B., Rees, D.C.(2001) Biochemistry 40: 641-650
- PubMed: 11170380
- DOI: https://doi.org/10.1021/bi001645e
- Primary Citation of Related Structures:
1G20, 1G21 - PubMed Abstract:
A mutant form of the nitrogenase iron protein with a deletion of residue Leu 127, located in the switch II region of the nucleotide binding site, forms a tight, inactive complex with the nitrogenase molybdenum iron (MoFe) protein in the absence of nucleotide. The structure of this complex generated with proteins from Azotobacter vinelandii (designated the L127Delta-Av2-Av1 complex) has been crystallographically determined in the absence of nucleotide at 2.2 A resolution and with bound MgATP (introduced by soaking) at 3.0 A resolution. As observed in the structure of the complex between the wild-type A. vinelandii nitrogenase proteins stabilized with ADP.AlF(4-), the most significant conformational changes in the L127Delta complex occur in the Fe-protein component. While the interactions at the interface between the MoFe-protein and Fe-proteins are conserved in the two complexes, significant differences are evident at the subunit-subunit interface of the dimeric Fe-proteins, with the L127Delta-Av2 structure having a more open conformation than the wild-type Av2 in the complex stabilized by ADP.AlF(4-). Addition of MgATP to the L127Delta-Av2-Av1 complex results in a further increase in the separation between Fe-protein subunits so that the structure more closely resembles that of the wild-type, nucleotide-free, uncomplexed Fe-protein, rather than the Fe-protein conformation in the ADP.AlF(4-) complex. The L127Delta mutation precludes key interactions between the Fe-protein and nucleotide, especially, but not exclusively, in the region corresponding to the switch II region of G-proteins, where the deletion constrains Gly 128 and Asp 129 from forming hydrogen bonds to the gamma-phosphate and activating water for attack on this group, respectively. These alterations account for the inability of this mutant to support mechanistically productive ATP hydrolysis. The ability of the L127Delta-Av2-Av1 complex to bind MgATP demonstrates that dissociation of the nitrogenase complex is not required for nucleotide binding.
- Division of Chemistry and Chemical Engineering, Mail Code 147-75CH, California Institute of Technology, Pasadena, California 91125, USA.
Organizational Affiliation: