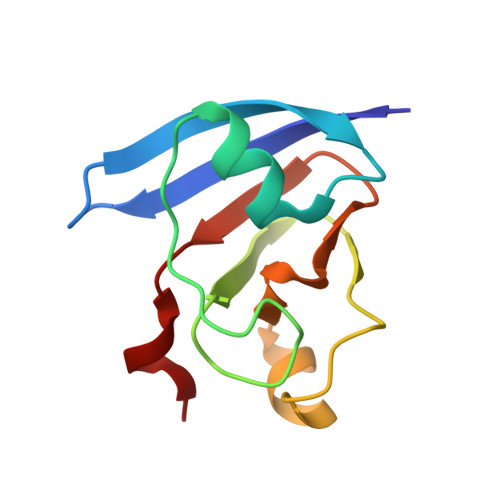
Molecular structure of the oxidized, recombinant, heterocyst [2Fe-2S] ferredoxin from Anabaena 7120 determined to 1.7-A resolution.
Jacobson, B.L., Chae, Y.K., Markley, J.L., Rayment, I., Holden, H.M.(1993) Biochemistry 32: 6788-6793
- PubMed: 8329401
- DOI: https://doi.org/10.1021/bi00077a033
- Primary Citation of Related Structures:
1FRD - PubMed Abstract:
The [2Fe-2S] ferredoxin produced in the heterocyst cells of Anabaena 7120 plays a key role in nitrogen fixation, where it serves as an electron acceptor from various sources and an electron donor to nitrogenase. The three-dimensional structure of this ferredoxin has now been determined and refined to a crystallographic R value of 16.7%, with all measured X-ray data from 30.0 to 1.7 A. The molecular motif of this ferredoxin is similar to that of other plant-type ferredoxins with the iron-sulfur cluster located toward the outer edge of the molecule and the irons tetrahedrally coordinated by both inorganic sulfurs and sulfurs provided by protein cysteinyl residues. The overall secondary structure of the molecule consists of seven strands of beta-pleated sheet, two alpha-helices, and seven type I turns. It is of special interest that 4 of the 22 amino acid positions thought to be absolutely conserved in nonhalophilic ferredoxins are different in the heterocyst form of the protein. Three of these positions are located in the metal-cluster binding loop.
- Institute for Enzyme Research, Graduate School, University of Wisconsin, Madison 53705.
Organizational Affiliation: