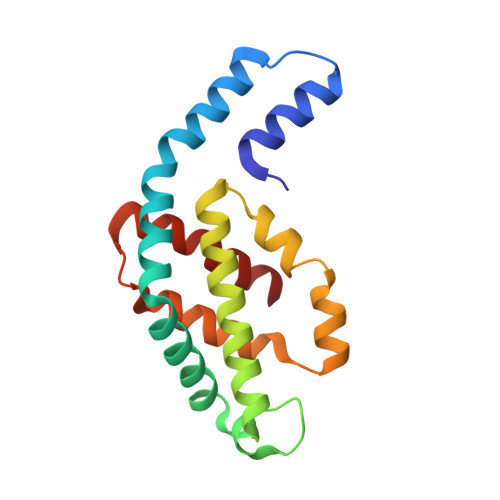
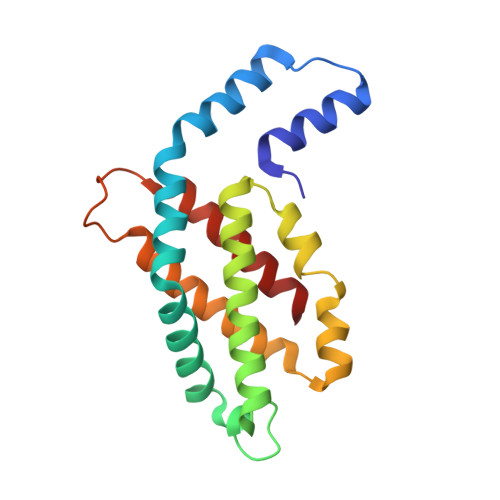
Crystal structure of R-phycocyanin and possible energy transfer pathways in the phycobilisome.
Jiang, T., Zhang, J.P., Chang, W.R., Liang, D.C.(2001) Biophys J 81: 1171-1179
- PubMed: 11463658
- DOI: https://doi.org/10.1016/S0006-3495(01)75774-8
- Primary Citation of Related Structures:
1F99 - PubMed Abstract:
The crystal structure of R-phycocyanin from Polysiphonia urceolata (R-PC-PU) at 2.4 A is reported. The R-PC-PU crystal belongs to space group P4(3)2(1)2 with cell parameters a = 135.1 A, c = 210.0 A, and alpha = beta = gamma = 90 degrees. The structure was determined by molecular replacement. The crystallographic R-factor of the refined model is 0.189 (R(free) = 0.239). Comparison of the microenvironment of chromophore beta 155 in R-PC-PU and in C-PC from Fremyolla diphosiphon (C-PC-FD) reveals that their spectral differences may be caused by their different alpha 28 residues. In the R-PC-PU crystal structure, two (alpha beta)(3) trimers assemble face to face to form a hexamer, and two such hexamers assemble in two novel side-to-side arrangements. Possible models for the energy transfer from phycoerythrin to phycocyanin and from phycocyanin to allophycocyanin are proposed based on several phycobiliprotein crystal structures.
- National Laboratory of Biomacromolecules, Institute of Biophysics, Chinese Academy of Sciences, Beijing 100101, China.
Organizational Affiliation: