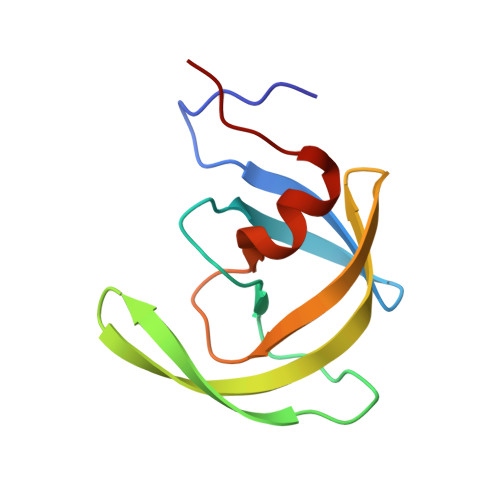
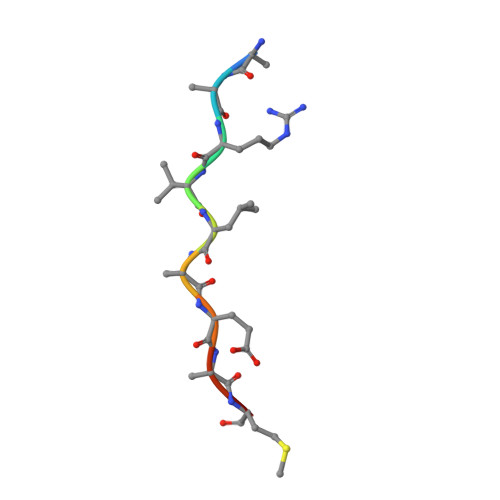
How does a symmetric dimer recognize an asymmetric substrate? A substrate complex of HIV-1 protease.
Prabu-Jeyabalan, M., Nalivaika, E., Schiffer, C.A.(2000) J Mol Biology 301: 1207-1220
- PubMed: 10966816
- DOI: https://doi.org/10.1006/jmbi.2000.4018
- Primary Citation of Related Structures:
1F7A - PubMed Abstract:
The crystal structure of an actual HIV-1 protease-substrate complex is presented at 2.0 A resolution (R-value of 19.7 % (R(free) 23.3 %)) between an inactive variant (D25N) of HIV-1 protease and a long substrate peptide, Lys-Ala-Arg-Val-Leu-Ala-Glu-Ala-Met-Ser, which covers a full binding epitope of capsid(CA)-p2, cleavage site. The substrate peptide is asymmetric in both size and charge distribution. To accommodate this asymmetry the two protease monomers adopt different conformations burying a total of 1038 A(2) of surface area at the protease-substrate interface. The specificity for the CA-p2 substrate peptide is mainly hydrophobic, as most of the hydrogen bonds are made with the backbone of the peptide substrate. Two water molecules bridge the two monomers through the loops Gly49-Gly52 (Gly49'-Gly52') and Pro79'-Val82' (Pro79-Val82). When other complexes are compared, the mobility of these loops is correlated with the content of the P1 and P1' sites. Interdependence of the conformational changes allows the protease to exhibit its wide range of substrate specificity.
- Department of Pharmacology and Molecular Toxicology, University of Massachusetts Medical School, Worcester, MA, 01655, USA.
Organizational Affiliation: