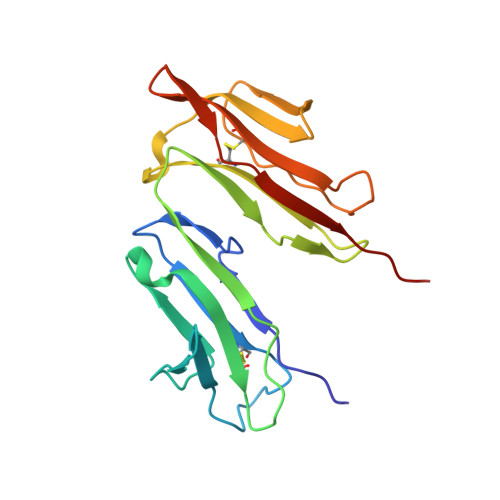
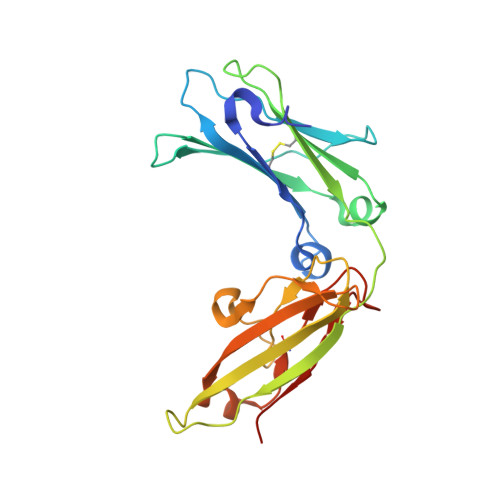
Structure of the Fc fragment of human IgE bound to its high-affinity receptor Fc (epsilon) RI (alpha).
Garman, S.C., Wurzburg, B.A., Tarchevskaya, S.S., Kinet, J.P., Jardetzky, T.S.(2000) Nature 406: 259-266
- PubMed: 10917520
- DOI: https://doi.org/10.1038/35018500
- Primary Citation of Related Structures:
1F6A - PubMed Abstract:
The initiation of immunoglobulin-E (IgE)-mediated allergic responses requires the binding of IgE antibody to its high-affinity receptor, Fc epsilonRI. Crosslinking of Fc epsilonRI initiates an intracellular signal transduction cascade that triggers the release of mediators of the allergic response. The interaction of the crystallizable fragment (Fc) of IgE (IgE-Fc) with Fc epsilonRI is a key recognition event of this process and involves the extracellular domains of the Fc epsilonRI alpha-chain. To understand the structural basis for this interaction, we have solved the crystal structure of the human IgE-Fc-Fc epsilonRI alpha complex to 3.5-A resolution. The crystal structure reveals that one receptor binds one dimeric IgE-Fc molecule asymmetrically through interactions at two sites, each involving one C epsilon3 domain of the IgE-Fc. The interaction of one receptor with the IgE-Fc blocks the binding of a second receptor, and features of this interaction are conserved in other members of the Fc receptor family. The structure suggests new approaches to inhibiting the binding of IgE to Fc epsilonRI for the treatment of allergy and asthma.
- Department of Biochemistry, Molecular Biology and Cell Biology, Northwestern University, Evanston, Illinois 60208, USA.
Organizational Affiliation: