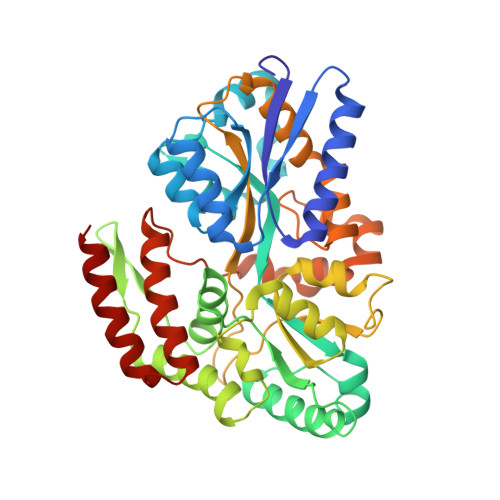
The crystal structure of a liganded trehalose/maltose-binding protein from the hyperthermophilic Archaeon Thermococcus litoralis at 1.85 A.
Diez, J., Diederichs, K., Greller, G., Horlacher, R., Boos, W., Welte, W.(2001) J Mol Biology 305: 905-915
- PubMed: 11162101
- DOI: https://doi.org/10.1006/jmbi.2000.4203
- Primary Citation of Related Structures:
1EU8 - PubMed Abstract:
We report the crystallization and structure determination at 1.85 A of the extracellular, membrane-anchored trehalose/maltose-binding protein (TMBP) in complex with its substrate trehalose. TMBP is the substrate recognition site of the high-affinity trehalose/maltose ABC transporter of the hyperthermophilic Archaeon Thermococcus litoralis. In vivo, this protein is anchored to the membrane, presumably via an N-terminal cysteine lipid modification. The crystallized protein was N-terminally truncated, resulting in a soluble protein exhibiting the same binding characteristics as the wild-type protein. The protein shows the characteristic features of a transport-related, substrate-binding protein and is structurally related to the maltose-binding protein (MBP) of Escherichia coli. It consists of two similar lobes, each formed by a parallel beta-sheet flanked by alpha-helices on both sides. Both are connected by a hinge region consisting of two antiparallel beta-strands and an alpha-helix. As in MBP, the substrate is bound in the cleft between the lobes by hydrogen bonds and hydrophobic interactions. However, compared to maltose binding in MBP, direct hydrogen bonding between the substrate and the protein prevails while apolar contacts are reduced. To elucidate factors contributing to thermostability, we compared TMBP with its mesophilic counterpart MBP and found differences known from similar investigations. Specifically, we find helices that are longer than their structurally equivalent counterparts, and fewer internal cavities.
- Department of Biology, University of Konstanz, 78457 Konstanz, Germany.
Organizational Affiliation: