Structural characteristics for biological activity of heat-stable enterotoxin produced by enterotoxigenic Escherichia coli: X-ray crystallography of weakly toxic and nontoxic analogs.
Sato, T., Ozaki, H., Hata, Y., Kitagawa, Y., Katsube, Y., Shimonishi, Y.(1994) Biochemistry 33: 8641-8650
- PubMed: 8038153
- DOI: https://doi.org/10.1021/bi00195a004
- Primary Citation of Related Structures:
1ETL, 1ETM - PubMed Abstract:
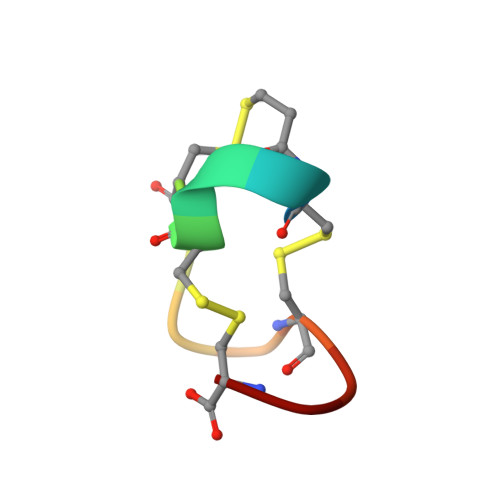
Heat-stable enterotoxin (ST) produced by a pathogenic strain of Escherichia coli exerts its function by binding to a membrane-bound guanylyl cyclase on intestinal epithelial cell membranes, which in turn catalyzes the production of cyclic GMP as a second messenger in the cells. To elucidate the structural requirements for the biological activities of ST, we synthesized [Mpr5,Gly13]STp(5-17) and [Mpr5,Leu13]STp(5-17), which are weakly toxic and nontoxic analogs of STp, in which the toxic domain consists of the sequence from Cys at position 5 to Cys at position 17. In these analogs, Cys at position 5 is replaced by Mpr (beta-mercaptopropionic acid) and Ala at position 13 by Gly and Leu, respectively. We examined these analogs by X-ray diffraction analysis using direct methods and refined the structures to crystallographic R factors of 7.3% and 6.6% using 5492 and 5122 data, respectively, observed > 3 sigma (Fo) with a resolution of 0.89 A. These peptides have a right-handed spiral structure consisting of three structural segments: an N-terminal 3(10) helix, a central type I beta-turn, and a C-terminal type II beta-turn. These structures show minor differences from that of [Mpr5]STp(5-17), the fully toxic analog of heat-stable enterotoxin [Ozaki et al. (1991) J. Biol. Chem. 266, 5934-5941], suggesting that the decrease and loss of the biological activities of [Mpr5,Gly13]STp(5-17) and [Mpr5,Leu13]STp(5-17), respectively, are not caused by structural changes but are associated with the direct interaction of Ala13 with the receptor protein. Careful comparison of these structures in crystalline states revealed that ST has the following structural characteristics: (i) inherent flexibility at the junctions of the three segments and in the central segment, which includes the putative receptor-binding residues, Ala13, (ii) a specific hydrophobic character around the central segment, and (iii) an unexpected C-terminal folding similar to those of functionally unrelated peptides that are known to be ionophores.
- Institute for Protein Research, Osaka University, Japan.
Organizational Affiliation: