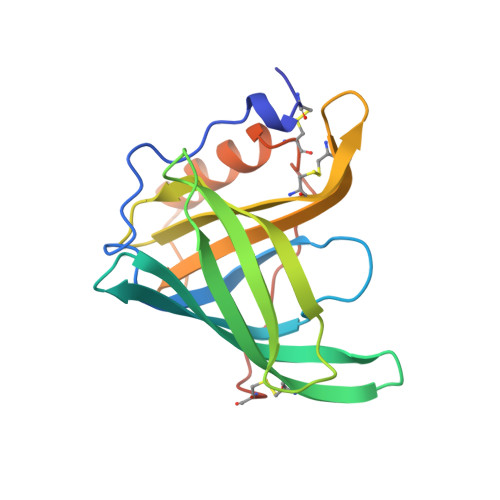
The interaction of N-ethyl retinamide with plasma retinol-binding protein (RBP) and the crystal structure of the retinoid-RBP complex at 1.9-A resolution.
Zanotti, G., Malpeli, G., Berni, R.(1993) J Biological Chem 268: 24873-24879
- PubMed: 8227049
- DOI: https://doi.org/10.2210/pdb1erb/pdb
- Primary Citation of Related Structures:
1ERB - PubMed Abstract:
The three-dimensional structure of bovine plasma retinol-binding protein (RBP) complexed with N-ethyl retinamide has been determined at 1.9-A resolution. The crystals of the complex (space group P2(1)2(1)2(1), a = 46.27, b = 49.11, c = 76.41 A) are isomorphous with those of bovine holo and apoRBP. The final crystallographic R factor is 0.172 for 11,261 observed reflections. The model of the retinoid-RBP complex is nearly identical to that of bovine retinol-RBP complex; the root mean square deviations between the alpha-carbons in the two proteins is 0.15 A. The N-ethyl retinamide molecule fits in the beta-barrel in the same position previously occupied by the vitamin. The ethyl group of the retinoid replaces the retinol hydroxyl group and a water molecule hydrogen bonded to it. This substitution has no consequence on the overall conformation of the protein. The modification of the functional end group of retinol does not lead to an apparent loss of affinity of the retinol analog for apoRBP, as established by means of fluorometric titrations with N-ethyl retinamide. However, the binding of the retinoid to RBP abolishes or greatly reduces the affinity of RBP for transthyretin. This behavior further supports the hypothesis that the area of the entrance of the beta-barrel, which includes the ethyl group of the retinoid bound to RBP, is involved in the interaction with transthyretin. Moreover, it indicates a high degree of complementarity of interacting surfaces between RBP and transthyretin. In fact, the replacement of the retinol hydroxyl group and quite small structural changes in the above region of the RBP molecule drastically affect the protein-protein recognition.
- Department of Organic Chemistry, University of Padova, Italy.
Organizational Affiliation: