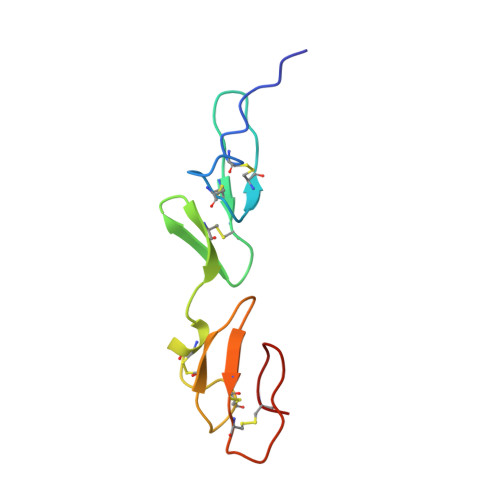
Solution structure of a pair of calcium-binding epidermal growth factor-like domains: implications for the Marfan syndrome and other genetic disorders.
Downing, A.K., Knott, V., Werner, J.M., Cardy, C.M., Campbell, I.D., Handford, P.A.(1996) Cell 85: 597-605
- PubMed: 8653794
- DOI: https://doi.org/10.1016/s0092-8674(00)81259-3
- Primary Citation of Related Structures:
1EMN, 1EMO - PubMed Abstract:
The nuclear magnetic resonance structure of a covalently linked pair of calcium-binding (cb) epidermal growth factor-like (EGF) domains from human fibrillin-1, the protein defective in the Marfan syndrome, is described. The two domains are in a rigid, rod-like arrangement, stabilized by interdomain calcium binding and hydrophobic interactions. We propose a model for the arrangement of fibrillin monomers in microfibrils that reconciles structural and antibody binding data, and we describe a set of disease-causing mutations that provide the first clues to the specificity of cbEFG interactions. The residues involved in stabilizing the domain linkage are highly conserved in fibrillin, fibulin, thrombomodulin, and the low density lipoprotein receptor. We propose that the relative orientation of tandem cbEGF domains in these proteins is similar, but that in others, including Notch, pairs adopt a completely different conformation.
- Department of Biochemistry University of Oxford, United Kingdom.
Organizational Affiliation: