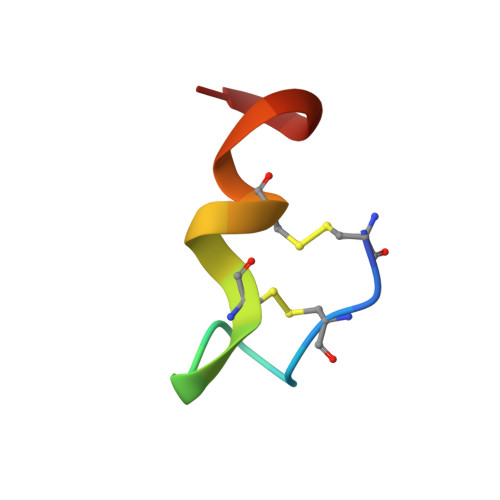
The crystal structure of human endothelin.
Janes, R.W., Peapus, D.H., Wallace, B.A.(1994) Nat Struct Biol 1: 311-319
- PubMed: 7664037
- DOI: https://doi.org/10.1038/nsb0594-311
- Primary Citation of Related Structures:
1EDN - PubMed Abstract:
The three-dimensional structure of the vasoactive polypeptide endothelin, the most potent vasoconstrictor yet identified, has been determined by X-ray crystallography to 2.18 A resolution. This intermediate-sized structure was solved by molecular replacement techniques using a fragment of an NMR-derived model for initial phasing of the data. However, comparisons of the final X-ray structure with the many diverse models derived from NMR data indicate some important differences, especially in the carboxy-terminal region of the molecule: the entire carboxy terminal tail (residues 16-21) is helical in the crystal structure, but not in any of the NMR structures. This may be a functionally significant difference as this region is crucial for receptor binding and vasoactivity.
- Department of Crystallography, Birkbeck College, University of London, UK.
Organizational Affiliation: