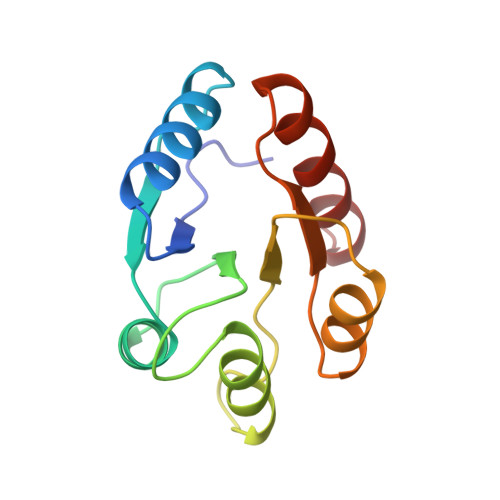
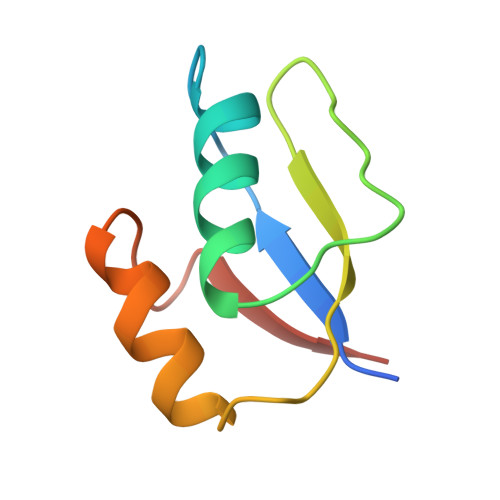
Two binding modes reveal flexibility in kinase/response regulator interactions in the bacterial chemotaxis pathway.
McEvoy, M.M., Hausrath, A.C., Randolph, G.B., Remington, S.J., Dahlquist, F.W.(1998) Proc Natl Acad Sci U S A 95: 7333-7338
- PubMed: 9636149
- DOI: https://doi.org/10.1073/pnas.95.13.7333
- Primary Citation of Related Structures:
1EAY - PubMed Abstract:
The crystal structure at 2.0-A resolution of the complex of the Escherichia coli chemotaxis response regulator CheY and the phosphoacceptor-binding domain (P2) of the kinase CheA is presented. The binding interface involves the fourth and fifth helices and fifth beta-strand of CheY and both helices of P2. Surprisingly, the two heterodimers in the asymmetric unit have two different binding modes involving the same interface, suggesting some flexibility in the binding regions. Significant conformational changes have occurred in CheY compared with previously determined unbound structures. The active site of CheY is exposed by the binding of the kinase domain, possibly to enhance phosphotransfer from CheA to CheY. The conformational changes upon complex formation as well as the observation that there are two different binding modes suggest that the plasticity of CheY is an essential feature of response regulator function.
- Institute of Molecular Biology, University of Oregon, Eugene, OR 97403, USA.
Organizational Affiliation: