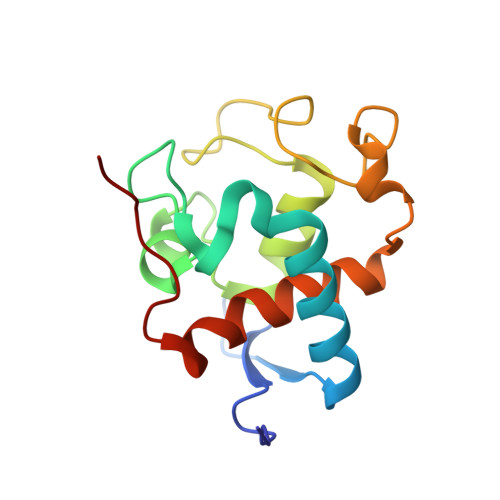
Crystal Structure of Low-Potential Cytochrome C549 from Synechocystis Sp. Pcc 6803 at 1.21A Resolution
Frazao, C., Enguita, F.J., Coelho, R., Sheldrick, G.M., Navarro, J.A., Hervas, M., De La Rosa, M.A., Carrondo, M.A.(2001) J Biol Inorg Chem 6: 324
- PubMed: 11315568
- DOI: https://doi.org/10.1007/s007750100208
- Primary Citation of Related Structures:
1E29 - PubMed Abstract:
The crystal structure of low-potential cytochrome c549, an extrinsic component of the photosystem II (PS II) from Synechocystis sp. PCC 6803, was obtained directly from single-wavelength 1.21 A resolution diffraction data. This is the first monodomain bis-histidinyl monoheme cytochrome c to be structurally characterized. The extended N-terminal region of c549 builds up a two-strand antiparallel beta-sheet in a hairpin motif, which extends through two molecules owing to crystal packing. Both peptide termini are involved in crystal contacts, which may explain their protrusion out of the globular fold. The C-terminus is preceded by a 9 A-long hydrophobic finger extending from a positively charged base and could be involved in PSII interactions, as well as a protruding negative patch built by a set of conserved acidic residues among c549 sequences.
- Instituto de Tecnologia Química e Biológica, Universidade Nova de Lisboa, 2781-901 Oeiras, Portugal.
Organizational Affiliation: