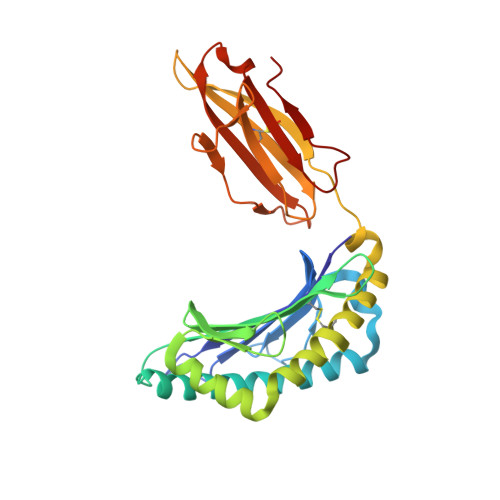
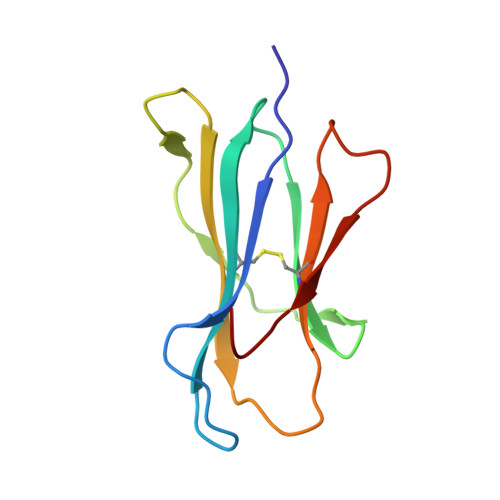
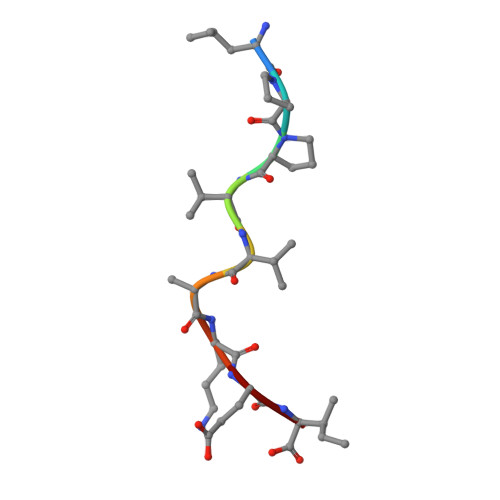
Nonstandard peptide binding revealed by crystal structures of HLA-B*5101 complexed with HIV immunodominant epitopes.
Maenaka, K., Maenaka, T., Tomiyama, H., Takiguchi, M., Stuart, D.I., Jones, E.Y.(2000) J Immunol 165: 3260-3267
- PubMed: 10975842
- DOI: https://doi.org/10.4049/jimmunol.165.6.3260
- Primary Citation of Related Structures:
1E27, 1E28 - PubMed Abstract:
The crystal structures of the human MHC class I allele HLA-B*5101 in complex with 8-mer, TAFTIPSI, and 9-mer, LPPVVAKEI, immunodominant peptide epitopes from HIV-1 have been determined by x-ray crystallography. In both complexes, the hydrogen-bonding network in the N-terminal anchor (P1) pocket is rearranged as a result of the replacement of the standard tyrosine with histidine at position 171. This results in a nonstandard positioning of the peptide N terminus, which is recognized by B*5101-restricted T cell clones. Unexpectedly, the P5 peptide residues appear to act as anchors, drawing the peptides unusually deeply into the peptide-binding groove of B51. The unique characteristics of P1 and P5 are likely to be responsible for the zig-zag conformation of the 9-mer peptide and the slow assembly of B*5101. A comparison of the surface characteristics in the alpha1-helix C-terminal region for B51 and other MHC class I alleles highlights mainly electrostatic differences that may be important in determining the specificity of human killer cell Ig-like receptor binding.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Headington, Oxford, United Kingdom. kmaenaka@lab.nig.ac.uk
Organizational Affiliation: