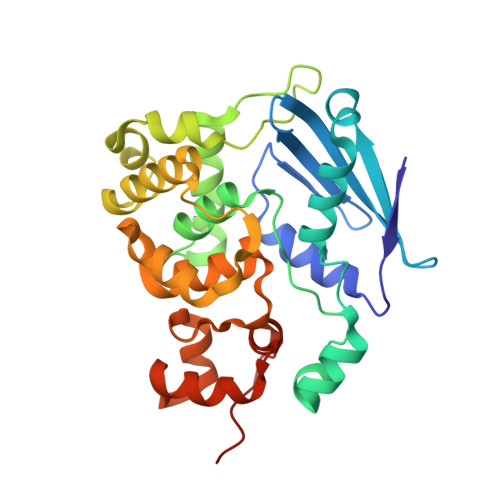
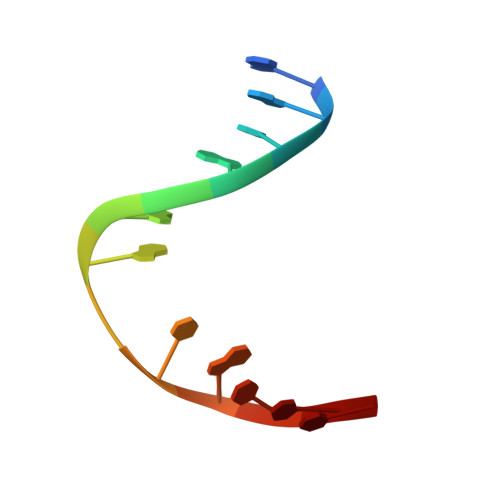
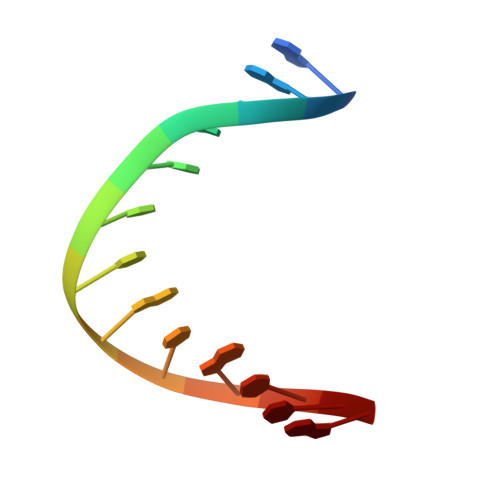
DNA bending and a flip-out mechanism for base excision by the helix-hairpin-helix DNA glycosylase, Escherichia coli AlkA.
Hollis, T., Ichikawa, Y., Ellenberger, T.(2000) EMBO J 19: 758-766
- PubMed: 10675345
- DOI: https://doi.org/10.1093/emboj/19.4.758
- Primary Citation of Related Structures:
1DIZ - PubMed Abstract:
The Escherichia coli AlkA protein is a base excision repair glycosylase that removes a variety of alkylated bases from DNA. The 2.5 A crystal structure of AlkA complexed to DNA shows a large distortion in the bound DNA. The enzyme flips a 1-azaribose abasic nucleotide out of DNA and induces a 66 degrees bend in the DNA with a marked widening of the minor groove. The position of the 1-azaribose in the enzyme active site suggests an S(N)1-type mechanism for the glycosylase reaction, in which the essential catalytic Asp238 provides direct assistance for base removal. Catalytic selectivity might result from the enhanced stacking of positively charged, alkylated bases against the aromatic side chain of Trp272 in conjunction with the relative ease of cleaving the weakened glycosylic bond of these modified nucleotides. The structure of the AlkA-DNA complex offers the first glimpse of a helix-hairpin-helix (HhH) glycosylase complexed to DNA. Modeling studies suggest that other HhH glycosylases can bind to DNA in a similar manner.
- Department of Biological Chemistry and Molecular Pharmacology, Harvard Medical School, 240 Longwood Avenue, Boston, MA 02115, USA.
Organizational Affiliation: