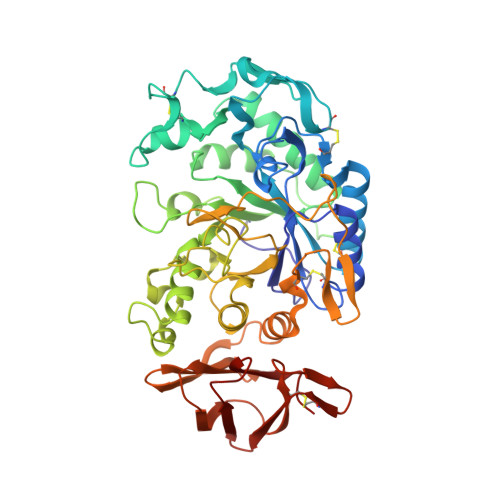
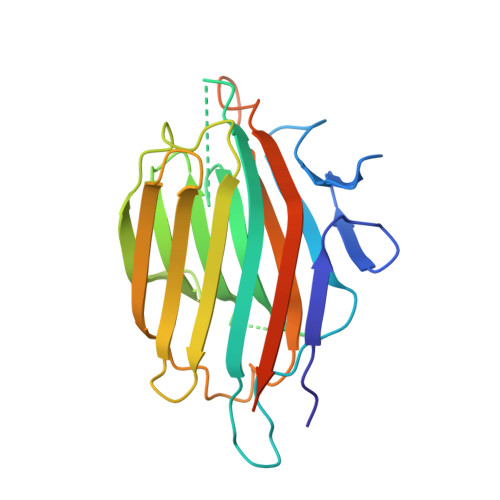
Substrate mimicry in the active center of a mammalian alpha-amylase: structural analysis of an enzyme-inhibitor complex.
Bompard-Gilles, C., Rousseau, P., Rouge, P., Payan, F.(1996) Structure 4: 1441-1452
- PubMed: 8994970
- DOI: https://doi.org/10.1016/s0969-2126(96)00151-7
- Primary Citation of Related Structures:
1DHK - PubMed Abstract:
alpha-Amylases catalyze the hydrolysis of glycosidic linkages in starch and other related polysaccharides. The alpha-amylase inhibitor (alpha-Al) from the bean Phaseolus vulgaris belongs to a family of plant defence proteins and is a potent inhibitor of mammalian alpha-amylases. The structure of pig pancreatic alpha-amylase (PPA) in complex with both a carbohydrate inhibitor (acarbose) and a proteinaceous inhibitor (Tendamistat) is known, but the catalytic mechanism is poorly understood. The crystal structure of pig pancreatic alpha-amylase complexed with alpha-Al was refined to 1.85 A resolution. It reveals that in complex with PPA, the inhibitor has the typical dimer structure common to legume lectins. Two hairpin loops extending out from the jellyroll fold of a monomer interact directly with the active site region of the enzyme molecule, with the inhibitor molecule filling the whole substrate-docking region of the PPA. The inhibitor makes substrate-mimetic interactions with binding subsites of the enzyme and targets catalytic residues in the active site. Binding of inhibitor induces structural changes at the active site of the enzyme. The present analysis reveals that there are extensive interactions between the inhibitor and residues that are highly conserved in the active site of alpha-amylases; alpha-Al1 inactivates PPA through elaborate blockage of substrate-binding sites. It provides a basis to design peptide analogue inhibitors. alpha-Amylase inhibition is of interest from several points of view, for example the treatment of diabetes and for crop protection.
- AFMB-IBSM-CNRS, Marseille, France.
Organizational Affiliation: