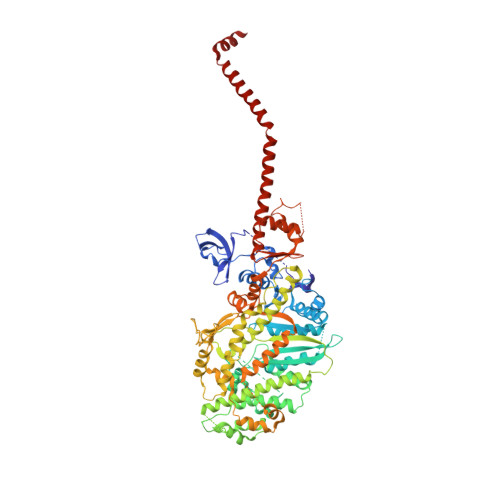
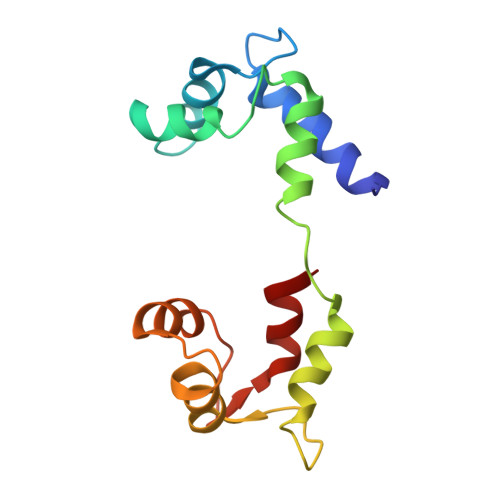
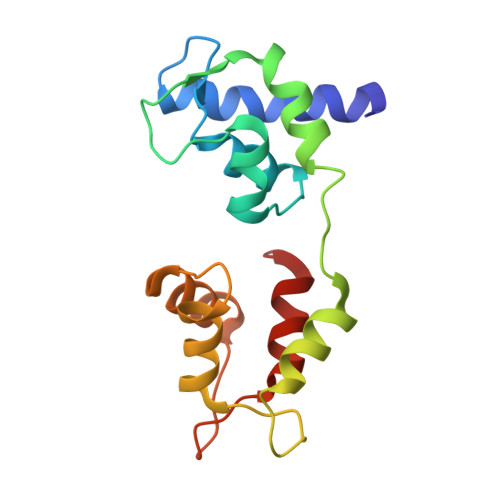
Three conformational states of scallop myosin S1.
Houdusse, A., Szent-Gyorgyi, A.G., Cohen, C.(2000) Proc Natl Acad Sci U S A 97: 11238-11243
- PubMed: 11016966
- DOI: https://doi.org/10.1073/pnas.200376897
- Primary Citation of Related Structures:
1DFK, 1DFL - PubMed Abstract:
We have determined the structure of the intact scallop myosin head, containing both the motor domain and the lever arm, in the nucleotide-free state and in the presence of MgADP.V04, corresponding to the transition state. These two new structures, together with the previously determined structure of scallop S1 complexed with MgADP (which we interpret as a detached ATP state), reveal three conformations of an intact S1 obtained from a single isoform. These studies, together with new crystallization results, show how the conformation of the motor depends on the nucleotide content of the active site. The resolution of the two new structures ( approximately 4 A) is sufficient to establish the relative positions of the subdomains and the overall conformation of the joints within the motor domain as well as the position of the lever arm. Comparison of available crystal structures from different myosin isoforms and truncated constructs in either the nucleotide-free or transition states indicates that the major features within the motor domain are relatively invariant in both these states. In contrast, the position of the lever arm varies significantly between different isoforms. These results indicate that the heavy-chain helix is pliant at the junction between the converter and the lever arm and that factors other than the precise position of the converter can influence the position of the lever arm. It is possible that this pliant junction in the myosin head contributes to the compliance known to be present in the crossbridge.
- Rosenstiel Basic Medical Sciences Research Center, and Biology Department, Brandeis University, Waltham, MA 02254-9110, USA. Anne.Houdusse@curie.fr
Organizational Affiliation: