Proteinase inhibitor homologues as potassium channel blockers.
Lancelin, J.M., Foray, M.F., Poncin, M., Hollecker, M., Marion, D.(1994) Nat Struct Biol 1: 246-250
- PubMed: 7544683
- DOI: https://doi.org/10.1038/nsb0494-246
- Primary Citation of Related Structures:
1DEM, 1DEN - PubMed Abstract:
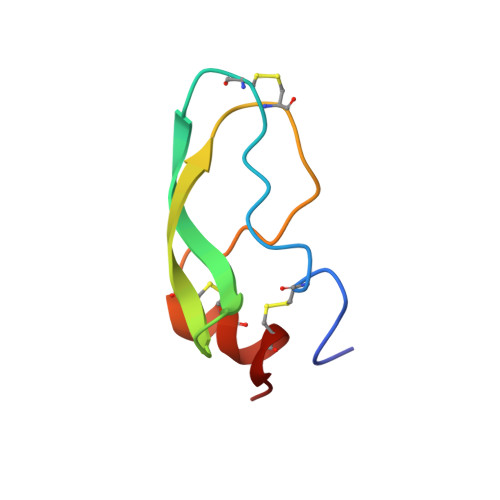
We report here the NMR structure of dendrotoxin I, a powerful potassium channel blocker from the venom of the African Elapidae snake Dendroaspis polylepis polylepis (black mamba), calculated from an experimentally-derived set of 719 geometric restraints. The backbone of the toxin superimposes on bovine pancreatic trypsin inhibitor (BPTI) with a root-mean-square deviation of < 1.7 A. The surface electrostatic potential calculated for dendrotoxin I and BPTI, reveal an important difference which might account for the differences in function of the two proteins. These proteins may provide examples of adaptation for specific and diverse biological functions while at the same time maintaining the overall three-dimensional structure of a common ancestor.
- Institute de Biologie Structurale CEA-CNRS, Grenoble, France.
Organizational Affiliation: