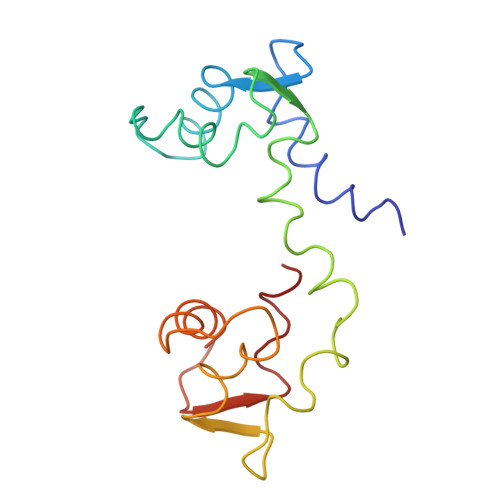
The linker of des-Glu84-calmodulin is bent.
Raghunathan, S., Chandross, R.J., Cheng, B.P., Persechini, A., Sobottka, S.E., Kretsinger, R.H.(1993) Proc Natl Acad Sci U S A 90: 6869-6873
- PubMed: 8341712
- DOI: https://doi.org/10.1073/pnas.90.14.6869
- Primary Citation of Related Structures:
1DEG - PubMed Abstract:
The crystal structure of a mutant calmodulin (CaM) lacking Glu-84 has been refined to R = 0.23 using data measured to 2.9-A resolution. In native CaM the central helix is fully extended, and the molecule is dumbbell shaped. In contrast, the deletion of Glu-84 causes a bend of 95 degrees in the linker region of the central helix at Ile-85. However, EF-hand domains 1 and 2 (lobe 1,2) do not touch lobe 3,4. The length, by alpha-carbon separation, of des-Glu84-CaM is 56 A; that of native CaM is 64 A. The shape of des-Glu84-CaM is similar to that of native CaM, as it is bound to the target peptide of myosin light-chain kinase. This result supports the proposal that the linker region of the central helix of CaM functions as a flexible tether.
- Department of Biology, University of Virginia, Charlottesville 22901.
Organizational Affiliation: